Team:Freiburg Transfection
From 2008.igem.org
(Difference between revisions)
| (One intermediate revision not shown) | |||
| Line 2: | Line 2: | ||
Content= | Content= | ||
<div style="font-size:18pt;"> | <div style="font-size:18pt;"> | ||
| - | <font face="Arial Rounded MT Bold" style="color:#010369">_transfection and synthetic receptor | + | <font face="Arial Rounded MT Bold" style="color:#010369">_transfection and synthetic receptor activation </font></div> |
<br><br> | <br><br> | ||
==Introduction== | ==Introduction== | ||
| Line 28: | Line 28: | ||
<br> | <br> | ||
[[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> | [[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> | ||
| - | ''Figure 1_Transfection: transfected 293T cells (left), untransfected cells (middle) and | + | ''Figure 1_Transfection: transfected 293T cells (left), untransfected cells (middle) and untransfected cells transmitted light (right)''<br> |
<br> | <br> | ||
<h3>'''Localization at the cell membrane'''</h3> | <h3>'''Localization at the cell membrane'''</h3> | ||
To show the localization of the constructs at the cell membrane transfection of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP was performed.<br> | To show the localization of the constructs at the cell membrane transfection of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP was performed.<br> | ||
| - | Figure 2_Transfection shows the | + | Figure 2_Transfection shows the model of the protein structure based on PDB files. Anti-Fluorescein Anticalin (Part BBa_K157004) is the extracellular part of the construct. The transmembrane region is identical to that of the EGF-receptor erbb1 (Part BBa_K157002). Split-beta-Lactamase, the intracellular part is labeled to the yellow fluorescent protein to detect membrane localization.<br> |
<br> | <br> | ||
[[Image:Freiburg2008_Lipo_bla1+YFP.jpg|500px]] | [[Image:Freiburg2008_Lipo_bla1+YFP.jpg|500px]] | ||
| Line 38: | Line 38: | ||
'''Figure 2_Transfection: Structure of the signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP construct. Extracellular: Lipocalin, GGGSlinker; Transmembrane: transmembraneregion of the EGF-receptor; Intracellular: Split-beta-Lactamase1, YFP.'''<br> | '''Figure 2_Transfection: Structure of the signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP construct. Extracellular: Lipocalin, GGGSlinker; Transmembrane: transmembraneregion of the EGF-receptor; Intracellular: Split-beta-Lactamase1, YFP.'''<br> | ||
<br> | <br> | ||
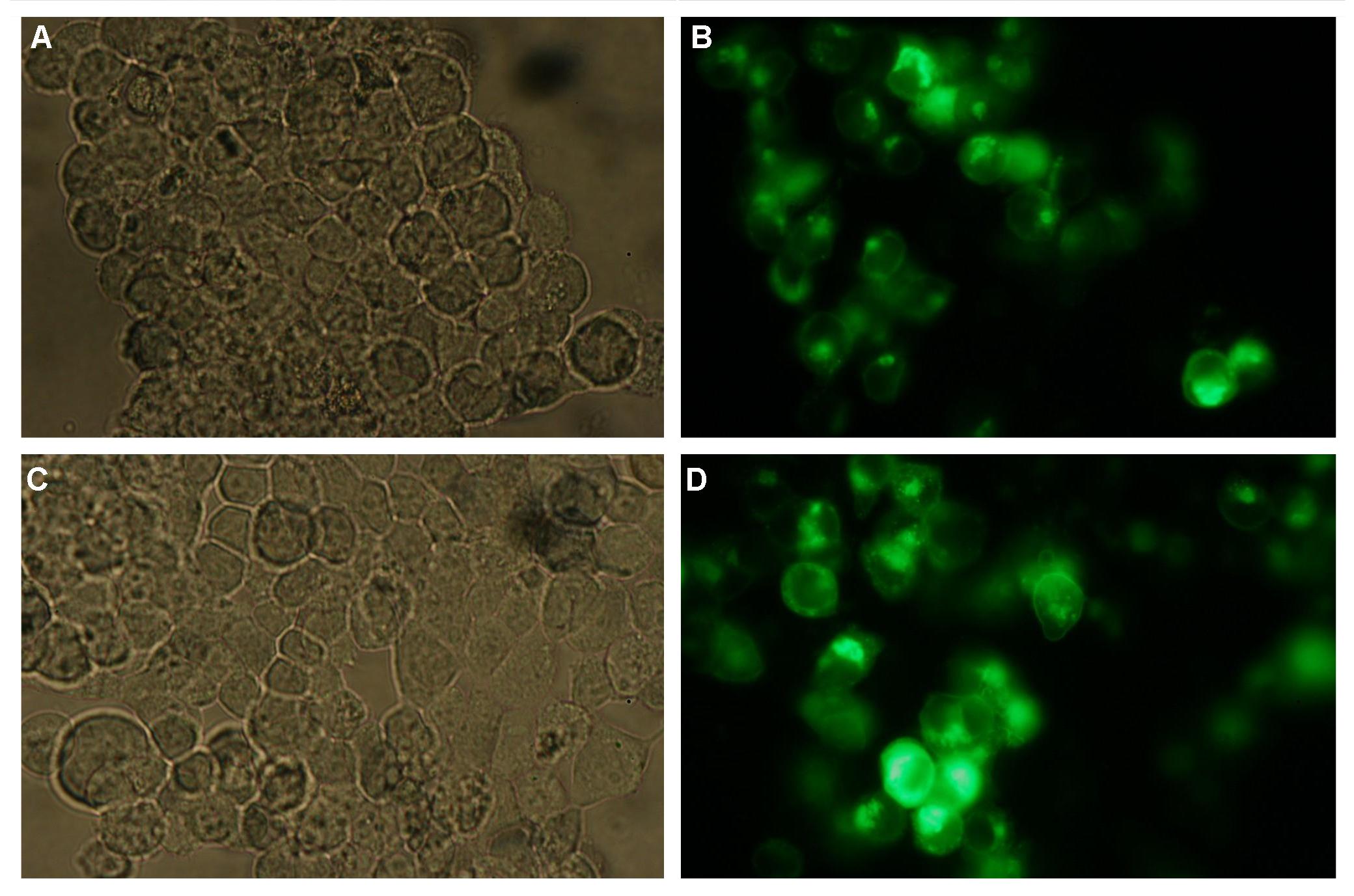
| - | Membranelocalization of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP is visible in transfected 293T cells (Figure 3_Transfection). The fluorescence of the cells is most likely restricted to the cellmembrane | + | Membranelocalization of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP is visible in transfected 293T cells (Figure 3_Transfection). The fluorescence of the cells is most likely restricted to the cellmembrane as evident by the strong signal at the surface of the cell spheres confirming the assembly of the construct in the cytoplasmamembrane.<br> |
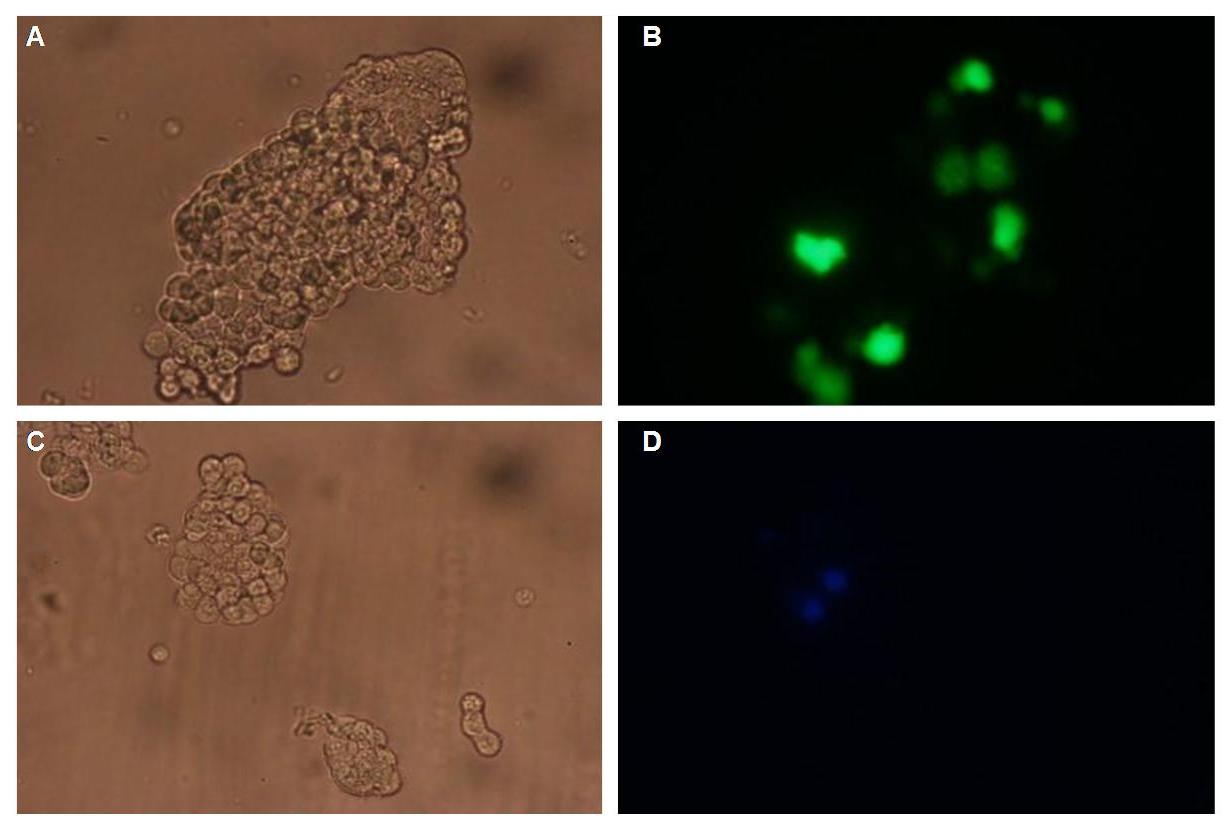
In comparison, 293T cells transfected with the construct transfectionvector-CMV-YFP show a uniformly distributed fluorescence all-over the cell (Figure 4_Transfection A and B).<br> | In comparison, 293T cells transfected with the construct transfectionvector-CMV-YFP show a uniformly distributed fluorescence all-over the cell (Figure 4_Transfection A and B).<br> | ||
| - | Transfection with the construct transfectionvector-CMV-CFP as well results in completely fluorescent cells (Figure 4_Transfection C and D).<br> | + | Transfection with the construct transfectionvector-CMV-CFP, i.e. without membrane targeting as well results in completely fluorescent cells (Figure 4_Transfection C and D).<br> |
<br> | <br> | ||
[[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP_1.jpg|710px]]<br> | [[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP_1.jpg|710px]]<br> | ||
| Line 54: | Line 54: | ||
'''Figure 5_Transfection: structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs'''<br> | '''Figure 5_Transfection: structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs'''<br> | ||
<br> | <br> | ||
| - | + | <h4>'''Outlook'''</h4> | |
| + | We hypothesize that adding fluorescein-coupled molecules leads to a clustering of the Lipocalin constructs due to the fluorescein-Lipocalin-binding. | ||
The clustering of the constructs in turn results in an assembly of the splitfluorophores or splitenzymes and therefore creates a functional protein (Figure 6_Transfection).<br> | The clustering of the constructs in turn results in an assembly of the splitfluorophores or splitenzymes and therefore creates a functional protein (Figure 6_Transfection).<br> | ||
<br> | <br> | ||
Latest revision as of 14:30, 29 October 2008
 "
"