Team:Chiba/Calendar-Home/30 August 2008
From 2008.igem.org
(→Meeting with UmeG) |
(→Team:Input) |
||
| (9 intermediate revisions not shown) | |||
| Line 16: | Line 16: | ||
x-RBS-cI-s-p(780bp) | x-RBS-cI-s-p(780bp) | ||
| - | |||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 65: | Line 64: | ||
-e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp) | -e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp) | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 103: | Line 102: | ||
-->'''[[Team:Chiba/protocol/DNA Purification/zymocleam|Zymo Clean]]''' | -->'''[[Team:Chiba/protocol/DNA Purification/zymocleam|Zymo Clean]]''' | ||
| - | eluted with | + | eluted with 10mL of NFW. |
-->'''[[Team:Chiba/protocol/ligation/dephosphorylation|SAP]]''' | -->'''[[Team:Chiba/protocol/ligation/dephosphorylation|SAP]]''' | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 144: | Line 143: | ||
'''[[Team:Chiba/protocol/ligation/ligation|Ligation]]''' | '''[[Team:Chiba/protocol/ligation/ligation|Ligation]]''' | ||
| - | + | ||
<table width="200" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="200" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 176: | Line 175: | ||
-->'''[[Team:Chiba/protocol/transformation|Transformation]]'''(XL10G) | -->'''[[Team:Chiba/protocol/transformation|Transformation]]'''(XL10G) | ||
| - | -->CFU40(b. | + | -->CFU40(b.g:22) |
'''[[Team:Chiba/protocol/PCR|PCR]]''' of 16 colonies) | '''[[Team:Chiba/protocol/PCR|PCR]]''' of 16 colonies) | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<tr> | <tr> | ||
| Line 216: | Line 215: | ||
</table> | </table> | ||
| - | -->[[Team:Chiba/protocol/gelcheck|Gel Check]] | + | -->[[Team:Chiba/protocol/gelcheck|Gel Check]] |
| - | result:One colony was apparent. | + | result:One colony was apparent. |
Picked the colony and incubate in 2mL of LB. | Picked the colony and incubate in 2mL of LB. | ||
| Line 230: | Line 229: | ||
x-RBS-cI-s-p(780bp) | x-RBS-cI-s-p(780bp) | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 266: | Line 265: | ||
'''[[Team:Chiba/protocol/ligation/gelex|Gel Extract]]''' | '''[[Team:Chiba/protocol/ligation/gelex|Gel Extract]]''' | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 291: | Line 290: | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 320: | Line 319: | ||
-e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp) | -e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp) | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 356: | Line 355: | ||
'''[[Team:Chiba/protocol/ligation/gelex|Gel Extract]]''' | '''[[Team:Chiba/protocol/ligation/gelex|Gel Extract]]''' | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 382: | Line 381: | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 407: | Line 406: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | -->left for an hour at 37 degrees,for 15 minutes at 65 degrees. | + | -->left for an hour at 37 degrees,for 15 minutes at 65 degrees. |
-->added 90μL Binding Buffer and vortexed. | -->added 90μL Binding Buffer and vortexed. | ||
| Line 418: | Line 417: | ||
| - | + | ||
<table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="150" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="57"></td> | <td width="57"></td> | ||
| Line 443: | Line 442: | ||
'''[[Team:Chiba/protocol/ligation/ligation|Ligation]]''' | '''[[Team:Chiba/protocol/ligation/ligation|Ligation]]''' | ||
| - | + | ||
<table width="200" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="200" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<td width="257"></td> | <td width="257"></td> | ||
| Line 472: | Line 471: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | -->left at rest at room temparature(25 degrees | + | -->left at rest at room temparature(25 degrees) |
-->'''[[Team:Chiba/protocol/transformation|Transformation]]'''(XL10G): | -->'''[[Team:Chiba/protocol/transformation|Transformation]]'''(XL10G): | ||
| - | -->CFU40(b. | + | -->CFU40(b.g:22) |
===Team:Communication=== | ===Team:Communication=== | ||
| Line 494: | Line 493: | ||
:(29/8)--->'''[[Team:Chiba/protocol/DNA Purification/sigma|Mini prep]]''' | :(29/8)--->'''[[Team:Chiba/protocol/DNA Purification/sigma|Mini prep]]''' | ||
| - | ::#insert: | + | ::#insert:[http://partsregistry.org/Part:BBa_C0170 BBa_C0170] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] |
| - | ::#insert: | + | ::#insert:[http://partsregistry.org/Part:BBa_C0178 BBa_C0178] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] |
:--->'''[[Team:Chiba/protocol/digestion|Digestion Test]]''' | :--->'''[[Team:Chiba/protocol/digestion|Digestion Test]]''' | ||
| - | :: | + | ::#insert:[http://partsregistry.org/Part:BBa_C0170 BBa_C0170] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] -> Sample No.1~4 |
| - | :: | + | ::#insert:[http://partsregistry.org/Part:BBa_C0178 BBa_C0178] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] -> Sample No.5~8 |
::*[http://partsregistry.org/Part:BBa_T9002 BBa_T9002](2007) -> Sample No.9 | ::*[http://partsregistry.org/Part:BBa_T9002 BBa_T9002](2007) -> Sample No.9 | ||
::*Single Digestion of Sample No.1~9 -> Sample No.10~18 | ::*Single Digestion of Sample No.1~9 -> Sample No.10~18 | ||
| Line 513: | Line 512: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>XbaI</td><td>0.1</td><td>0.1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>SpeI</td><td>-</td><td>0.1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 528: | Line 527: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td>TOTAL</td><td> | + | <td>TOTAL</td><td>10μl</td><td>10μl</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 556: | Line 555: | ||
<tr> | <tr> | ||
<td>TOTAL</td> | <td>TOTAL</td> | ||
| - | <td> | + | <td>6μl</td><td>12μl</td><td>12μl</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 582: | Line 581: | ||
::23 -> OK?? | ::23 -> OK?? | ||
|} | |} | ||
| - | |||
===Team:Output=== | ===Team:Output=== | ||
| Line 592: | Line 590: | ||
[[Team:Chiba/protocol/PCR|PCR]] | [[Team:Chiba/protocol/PCR|PCR]] | ||
| - | *[http://partsregistry.org/Part:BBa_J52008 BBa_J52008] | + | *[http://partsregistry.org/Part:BBa_J52008 BBa_J52008](1) |
<table width="250" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | <table width="250" border="4" cellpadding="0" cellspacing="0" bordercolor="#000000"> | ||
<tr> | <tr> | ||
<td width="257">Sample No.</td> | <td width="257">Sample No.</td> | ||
| - | <td> | + | <td>1</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
Latest revision as of 23:13, 29 October 2008
29 August 2008 <|> 31 August 2008
Contents |
Meeting with UmeG
the ppt (in Japanese) for our weekly meeting
Laboratory work
Team:Input
x-RBS-cI-s-p(780bp)
| double | |
| dH2O(μL) | 6 |
| 10×BSA(μL) | 10 |
| 10×NE(μL) | 10 |
| PstI(μL) | 2 |
| XbaI(μL) | 2 |
| DNA(μL) | 33.6ng/μl×30μl(1μg) |
| TOTAL(μL) | 60 |
-->30μlGel extraction
-->Zymo Clean
eluted with 5μL of Nuclease free water(NFW)
Gel Check(1μl)
-->OK
(remains of the solution(4μL)-->Ligation)
-e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp)
| double | |
| dH2O(μL) | 4 |
| 10×BSA(μL) | 10 |
| 10×NE(μL) | 10 |
| PstI(μL) | 2 |
| SpeI(μL) | 4 |
| DNA(μL) | 75ng/μl×30μl(2μg) |
| TOTAL(μL) | 60 |
-->30μlGel extraction
-->Zymo Clean
eluted with 10mL of NFW.
-->SAP
| dH2O(μL) | 9 |
| DNA(μL) | 10 |
| SAP(μL) | 1 |
| SAP Buffer(μL) | 10 |
| TOTAL(μL) | 30 |
eluted with 5μL of Nuclease free water(NFW)
Gel Check(1μl)
-->OK
(remains of the solution(4μL)-->Ligation)
| - | b.g | |
| dH2O(μL) | 1.4 | 8 |
| Ligase(μL) | 1 | 1 |
| Ligase Buffer(μL) | 2 | 2 |
| Vector(μL) | 1.8 | 1 |
| Insert(μL) | 3 | - |
| TOTAL(μL) | 10 | 10 |
-->Transformation(XL10G)
-->CFU40(b.g:22)
PCR of 16 colonies)
| VF(μL) | 2 |
| VR2(μL) | 2 |
| dNTP(μL) | 2 |
| thermo pol buffer(μL) | 2 |
| Tag(μL) | 0.3 |
| dH2O(μL) | 11.7 |
| TOTAL(μL) | 20 |
-->Gel Check
result:One colony was apparent.
Picked the colony and incubate in 2mL of LB.
-->Mini prepped
x-RBS-cI-s-p(780bp)
| double | |
| dH2O(μL) | 6 |
| 10×BSA(μL) | 10 |
| 10×NE(μL) | 10 |
| PstI(μL) | 2 |
| XbaI(μL) | 2 |
| DNA(μL) | 33.6ng/μl×30μl(1μg) |
| TOTAL(μL) | 60 |
| Digested DNA Sample(μL) | 30 |
| Loading Dye(μL) | 6 |
| TOTAL(μL) | 36 |
-->Zymo Clean
-->eluted with 5μl of NFW
-->Gel Check
| dH2O(μL) | 4 |
| DNA(μL) | 1 |
| Loading Dye(μL) | 1 |
| TOTAL(μL) | 6 |
-->Gel Check-->OK
-e-x-Ptet-s-p-(PMB1,Amp)(Insert:54bp,Vector:2079bp)
| double | |
| dH2O(μL) | 4 |
| 10×BSA(μL) | 10 |
| 10×NE(μL) | 10 |
| PstI(μL) | 2 |
| SpeI(μL) | 4 |
| DNA(μL) | 75ng/μl×30μl(2.25μg) |
| TOTAL(μL) | 60 |
| Digested DNA Sample(μL) | 30 |
| Loading Dye(μL) | 6 |
| TOTAL(μL) | 36 |
-->Zymo Clean
-->eluted with 10μl of NFW
-->SAP
| dH2O(μL) | 9 |
| DNA(μL) | 10 |
| SAP(μL) | 1 |
| SAP Buffer(×10)(μL) | 10 |
| TOTAL(μL) | 30 |
-->left for an hour at 37 degrees,for 15 minutes at 65 degrees.
-->added 90μL Binding Buffer and vortexed.
-->Zymo Clean
-->eluted with 5μl of NFW
| dH2O(μL) | 4 |
| DNA(μL) | 1 |
| Loading Dye(μL) | 1 |
| TOTAL(μL) | 6 |
-->Gel Check-->OK
| - | b.g | |
| dH2O(μL) | 1.4 | 8 |
| Ligase(μL) | 1 | 1 |
| Ligase Buffer(μL) | 2 | 2 |
| Vector(μL) | 1.8 | 1 |
| Insert(μL) | 3 | - |
| TOTAL(μL) | 10 | 10 |
-->left at rest at room temparature(25 degrees) -->Transformation(XL10G):
-->CFU40(b.g:22)
Team:Communication
- Transformation
- competent cells : XL10G
- [http://partsregistry.org/Part:BBa_I9026 BBa_I9026](2007)
- [http://partsregistry.org/Part:BBa_I9030 BBa_I9030](2006)
- [http://partsregistry.org/Part:BBa_S03154 BBa_S03154](2007)
- [http://partsregistry.org/Part:BBa_S03156 BBa_S03156](2007)
- [http://partsregistry.org/Part:BBa_S03158 BBa_S03158](2007)
- [http://partsregistry.org/Part:BBa_S03160 BBa_S03160](2007)
- [http://partsregistry.org/Part:BBa_C0062 BBa_C0062](2007)
- [http://partsregistry.org/Part:BBa_C0179 BBa_C0179](2007)
- --->(31/8)Mini prep
- (29/8)--->Mini prep
- insert:[http://partsregistry.org/Part:BBa_C0170 BBa_C0170] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500]
- insert:[http://partsregistry.org/Part:BBa_C0178 BBa_C0178] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500]
- --->Digestion Test
- insert:[http://partsregistry.org/Part:BBa_C0170 BBa_C0170] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] -> Sample No.1~4
- insert:[http://partsregistry.org/Part:BBa_C0178 BBa_C0178] + vector:[http://partsregistry.org/Part:BBa_J04500 BBa_J04500] -> Sample No.5~8
- [http://partsregistry.org/Part:BBa_T9002 BBa_T9002](2007) -> Sample No.9
- Single Digestion of Sample No.1~9 -> Sample No.10~18
- Double Digestion of Sample No.1~9 -> Sample No.19~27
Sample No. Single : 10~18 Double : 19~27 Sample DNA 1 5 XbaI 0.1 0.1 SpeI - 0.1 Buffer 2 1 1 BSA 1 1 dH2O 6.9 2.8 TOTAL 10μl 10μl
- --->Gel Check
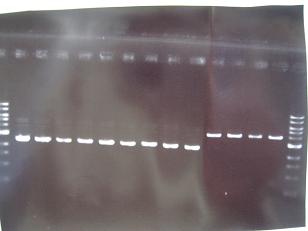
Sample No. 1~9 10~18 19~27 Sample DNA 1 10 10 Loading Dye 1 2 2 dH2O 4 - - TOTAL 6μl 12μl 12μl - From left;
- Sanple No.1~13
- -> OK
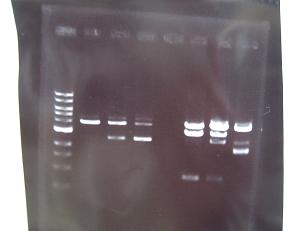
- From left;
- From left;
- Sample No.14~17,24~16
- 14 -> OK
- 15,16 -> Bad
- 17 -> None
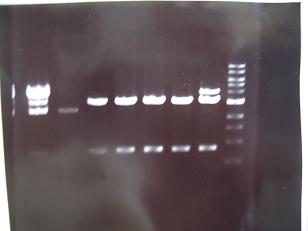
- 24~16 -> Bad
- From left;
- Sample No.18,27,19~23
- 18,27 -> Bad
- 19~22 -> OK
- 23 -> OK??
Team:Output
- mcherry
- [http://partsregistry.org/Part:BBa_J52008 BBa_J52008](1)
Sample No. 1 DNA tamplate 1 rLuc_fwd 2.5 Rev primer 2.5 Thermo pol Buffer 5 dNTPmix 5 Vent pol 0.5 dH2O 34 TOTAL 50μl
 "
"