Team:Newcastle University/Notebook
From 2008.igem.org
MarkWappett (Talk | contribs) |
|||
| (272 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Newcastle University/Header}} | |
| - | + | {{:Team:Newcastle University/Template:UnderTheHome|page-title=[[Team:Newcastle University/Notebook|Wet Lab]]}} | |
| - | + | {{:Team:Newcastle University/WetlabSideBar}} | |
| + | <div id="maincol"> | ||
| - | + | ==Introduction== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The overall design was too complicated for the time and resources available so we devised a proof of concept Brick which represented an input node from our full design. This Brick demonstrates that it is possible to transfer two-component peptide sensing from one strain into another and have it function as it did in the first strain. We took the subtilin two component system from ''Bacillus subtilis'' ATCC6633 and integrated it into the chromosome of ''B. subtilis'' 168, the typical laboratory ''B. subtilis'' strain at the ''amyE'' locus. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The wet lab work involved: | |
| - | == | + | * Cloning the synthesized Brick from pUC57 in ''E. coli'' into our ''Bacillus'' integration vectors (with GFP and mCherry respectively downstream of the Brick) |
| + | * Validation of the integration vector | ||
| + | * Transformation and integration into the chromosome of ''B. subtilis'' 168 at the ''amyE'' locus | ||
| + | * Validation of chromosomal integration | ||
| + | * Characterisation of the integrated parts and their response to subtilin. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | Our subtilin sensor, part [http://partsregistry.org/Part:BBa_K104001 BBa_k104001] | ||
| + | |||
| + | [[Image:biobrickk.gif|thumb|center|300px|Figure 1: Newcastle's iGEM team construct.]] | ||
| + | |||
| + | Figure 1 shows the construct which contains: | ||
| + | * ''spaRK'' promotor | ||
| + | * ''spaR'' (subtilin peptide antibiotic Regulation) - the 220 amino acid product of this gene usually regulates the downstream production of subtilin antibiotic. It has an N-terminal domain that can be phosphorylated and a C-terminal domian that has DNA binding properties [http://aem.asm.org/cgi/reprint/59/1/296.pdf] | ||
| + | * ''spaK'' (subtilin peptide antibiotic Kinase) - this gene codes for a 325 amino acid histadine kinase peptide that phosphorylates the N-terminus of ''spaR'' [http://aem.asm.org/cgi/reprint/59/1/296.pdf]. This activates the DNA binding ability of the C-terminus of ''spaR'', which in turn initiates transcription of the downstream gene. In the case of our construct, this gene is ''gfp''. | ||
| + | * ''rrnO'' transcriptional terminator | ||
| + | * ''spaS'' promotor - a strong promotor inducible by upstream activation of ''spaRK''. It can be placed in front any gene to regulate its activity. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | '''Aim 1: clone the ''spaRK'' system from pUC57 into pJWV021 and transform into DH5 alpha competent ''E. coli''''' | ||
| + | |||
| + | |||
| + | [[Image:cloning 1.jpg|thumb|center|300px|Aim 1]] | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | '''Aim 2: clone the ''spaRK'' system from pUC57 into pGFP-rrnB and transform into DH5 alpha competent ''E. coli''''' | ||
| + | |||
| + | |||
| + | [[Image:cloning 2.jpg|thumb|center|300px|Aim 2]] | ||
| + | |||
| + | * The 2.2kb fragment (ncl08) contains the ''spaRK'' biobrick system. When properly cloned, our biobrick will replace the rrnB promoter in front of gfp present in plasmid pGFP-rrnB. Thus, gfp will now be under control of the subtilin inducible spaS promoter. This means that when ''SpaR'' is activated by SpaK (by sensing subtilin) its positive regulatory effect on PspaS will activate ''gfp''. | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | '''Aim 3: clone the ''agr'' (Cambridge biobrick for sensing S. aureus) system from the biobrick plasmid using EcoRI and SpeI sites into the corresponding sites of pGFP-rrnB. This will replace the PrrnB-gfp on the bacillus vector with the agr biobrick. The ligation was transformed into DH5 alpha competent ''E. coli''''' | ||
| + | |||
| + | |||
| + | [[Image:cloning 3.jpg|thumb|center|300px|Aim 3]] | ||
| + | |||
| + | * BBa_I746107 contains a partial ''agr'' operon, which includes ''agrC'' and ''A'' but has ''agrB'' and ''D'' deleted. This allows coding for the receptor (''C''/''A'') but not for production of the quorum peptides themselves (''B''/''D''). The inducible promoter drives expression of gfp. | ||
| + | |||
| + | |||
| + | |||
| + | <html><a name="subcloning"></html> | ||
| + | == Subcloning == | ||
| + | |||
| + | The pUC57 was restriction-digested and the fragments displayed on a gel to ensure that the correct products have been produced. | ||
| + | |||
| + | [[Image:02091.jpg|500px]] | ||
| + | Figure 2: Gel from [[Newcastle_University_Wetlab/2_September_2008|2 Sept]] showing our 3 unrestricted plasmids, along with the 4 restrictions | ||
| + | * '''Lane 1:''' 1kb ladder | ||
| + | * '''Lane 2:''' Unrestricted pGFP-rrnB sample 1 (our bacillus integration vector that carries gfp between 2 amyE flanking regions) | ||
| + | * '''Lane 3:''' Unrestricted pGFP-rrnB sample 2 | ||
| + | * '''Lane 4:''' Unrestricted pJWV021 sample 1 (our bacillus integration vector that carries mCherry between 2 sacA flanking regions) | ||
| + | |||
| + | * '''Lane 5:''' Unrestricted pJWV021 sample 2 | ||
| + | * '''Lane 6:''' Unrestricted pUC57-ncl08 sample 1 (our synthetic biobrick construct; GenScript) | ||
| + | * '''Lane 7:''' Unrestricted pUC57-ncl08 sample 2 | ||
| + | * '''Lane 8:''' 1kb ladder | ||
| + | * '''Lane 9:''' pGFP-rrnB restricted with EcoR1 and Nhe1 | ||
| + | * '''Lane 10:''' pJWV021 restricted with BglII and Nhe1 | ||
| + | * '''Lane 11:''' pUC57-ncl08 restricted with EcoR1 and Nhe1 | ||
| + | * '''Lane 12:''' pUC57-ncl08 restricted with BglII and Nhe1 | ||
| + | |||
| + | Not shown here is the agr biobrick digestion. | ||
| + | |||
| + | <html><a name="vector-validation"></html> | ||
| + | == Validation of the integration vector == | ||
| + | |||
| + | |||
| + | Digested pGFP-rrnB/pJWV021 were mixed with digested pUC57-ncl08 or agr biobrick, ligated and transformed to E.coli and the bacteria [[Newcastle_University_Wetlab/4_September_2008|overnight]] were grown on selective media (the pGFP ligation on plates with spectinomycin (50microG/ml) and the pJWV021 ligation on kanamycin (10 microG/ml)). Transformants were grown up in LB with antibiotics, plasmid was isolated and digested to check for successful subcloning (in the GFP and mCherry vectors, GFP results shown below). | ||
| + | |||
| + | [[Image:Gel_2.jpg|500px]] | ||
| + | Figure 4: Gel showing restricted pGFPrrnB-ncl08 taken from 12 different colonies | ||
| + | * '''Lane 1:''' 1kb ladder | ||
| + | * '''Lane 2:''' pGFPrrnB-ncl08 colony 1 (white colony, 25μl plate) | ||
| + | * '''Lane 3:''' pGFPrrnB-ncl08 colony 2 (white colony, 25μl plate) | ||
| + | * '''Lane 4:''' pGFPrrnB-ncl08 colony 3 (white colony, 25μl plate) | ||
| + | * '''Lane 5:''' pGFPrrnB-ncl08 colony 4 (white colony, 25μl plate) | ||
| + | * '''Lane 6:''' pGFPrrnB-ncl08 colony 5 (white colony, 25μl plate) | ||
| + | * '''Lane 7:''' pGFPrrnB-ncl08 colony 6 (white colony, 25μl plate) | ||
| + | * '''Lane 8:''' pGFPrrnB-ncl08 colony 7 (white colony, 25μl plate) | ||
| + | * '''Lane 9:''' pGFPrrnB-ncl08 colony 8 (white colony, 25μl plate) | ||
| + | * '''Lane 10:''' pGFPrrnB-ncl08 colony 9 (white colony, 25μl plate) | ||
| + | * '''Lane 11:''' pGFPrrnB-ncl08 colony 10 (white colony, 250μl plate) | ||
| + | * '''Lane 12:''' pGFPrrnB-ncl08 colony 11 (white colony, 250μl plate) | ||
| + | * '''Lane 13:''' pGFPrrnB-ncl08 colony 12 (green colony, 25μl plate) | ||
| + | |||
| + | This demonstrated that we had succesfully subcloned our ncl08 biobrick from pUC57-ncl08 to pGFP. Not shown here are the restriction checks for pJWV021-ncl08 and pGFP-agr (but we have correct clones for them as well!). | ||
| + | |||
| + | |||
| + | <html><a name="bsubtilis-integration"></html> | ||
| + | ==Transformation and integration into the ''B. subtilis'' chromosome== | ||
| + | |||
| + | [[Image:CloningStratStep2.jpg|500px|thumb|left]] | ||
| + | |||
| + | |||
| + | <html><a name="chromosome-validation"></html> | ||
| + | ==Validation of chromosomal integration== | ||
| + | |||
| + | Next, we transformed the correct plasmids to (self made) competent ''Bacillus'', as described in the protocol section. For plasmid pGFP-ncl08 and pGFP-agr, bacillus was plated on chloramphenicl (5 microG/ml) and for plasmid pJWV021-ncl08, bacillus were plated on kanamycin (5 microG/ml). Next day, Bacillus transformants were subseqently streaked to single colonies on selective agar plates. To check for correct integration of the pGFP-ncl08 and pGFP-agr plasmids at the amyE locus, single colonies were streaked on nutrient agar plates containing 1% starch. If a double crossover event occured, cells are not able to produce AmyE and will not degrade starch, thus no 'halo' around the colony. 2 individual colonies without an halo were grown and stocked and used for later analyses. To check correct integration of the pJWV021-ncl08 plasmid in the Bacillus chromosome, single colonies were streaked on minimal medium plates containing either glucose or sucrose as the sole carbon source. If the plasmid integrated correctly at the sacA locus via a double crossover, cells cannot utilise sucrose anymore and will not grow on the minimal plate containing sucrose, but will grow on the glucose plate. Again, 2 correct transformants were selected that were unable to proliferate on sucrose plates and stocked in the -80C freezer for future work. | ||
| + | |||
| + | |||
| + | |||
| + | <html><a name="characterisation"></html> | ||
| + | ==Characterisation of the integrated parts and their response to subtilin== | ||
| + | |||
| + | <html><a name="results"></html> | ||
| + | == Results == | ||
| + | |||
| + | To analyse the results of the wet lab transformations of the inserts into ''B. subtilis'', we used two methods: microscopy and flow cytometry. | ||
| + | |||
| + | ===Microscopy=== | ||
| + | |||
| + | Microscopy work from 08.09.08 showed a difference in the level of flourescence of the iGEMgfp fluorescent cells (higher in 10% subtilin-induced cells compared to 0% subtilin-induced cells). However, there was little difference in the ''number'' of cells that fluoresced between the two cultures. | ||
| + | |||
| + | There was no difference in the number of fluorecent cells ''or'' the level of fluorescence between the 10% subtilin-induced and the 0% subtilin-induced iGEMcherry cells. | ||
| + | |||
| + | |||
| + | ===Flow cytometry=== | ||
| + | |||
| + | Flow cytometry measures cell fluorescence and light scattering, cell-by-cell, allowing us to quantify our results and present them in graphical form. A sample of our cells engineered ''B. subtilis'' cells were injected into the machine, which hydro-dynamically focusses the fluid. Lasers of a particular wavelength are directed onto the stream of fluid, and each particle which passes through the light beam will cause the laser to scatter in a different way. Cells which absorb and then re-emit the light, result in fluorescence (a higher energy state). | ||
| + | |||
| + | The detectors in the machine measure the scattering of light and any fluoresence which occurs, and the data is displayed as histograms and dot plots. | ||
| + | |||
| + | '''Induction by 0% v/v subtilin''' (i.e in the absence of subtilin): mean fluoresence = 7.70 | ||
| + | |||
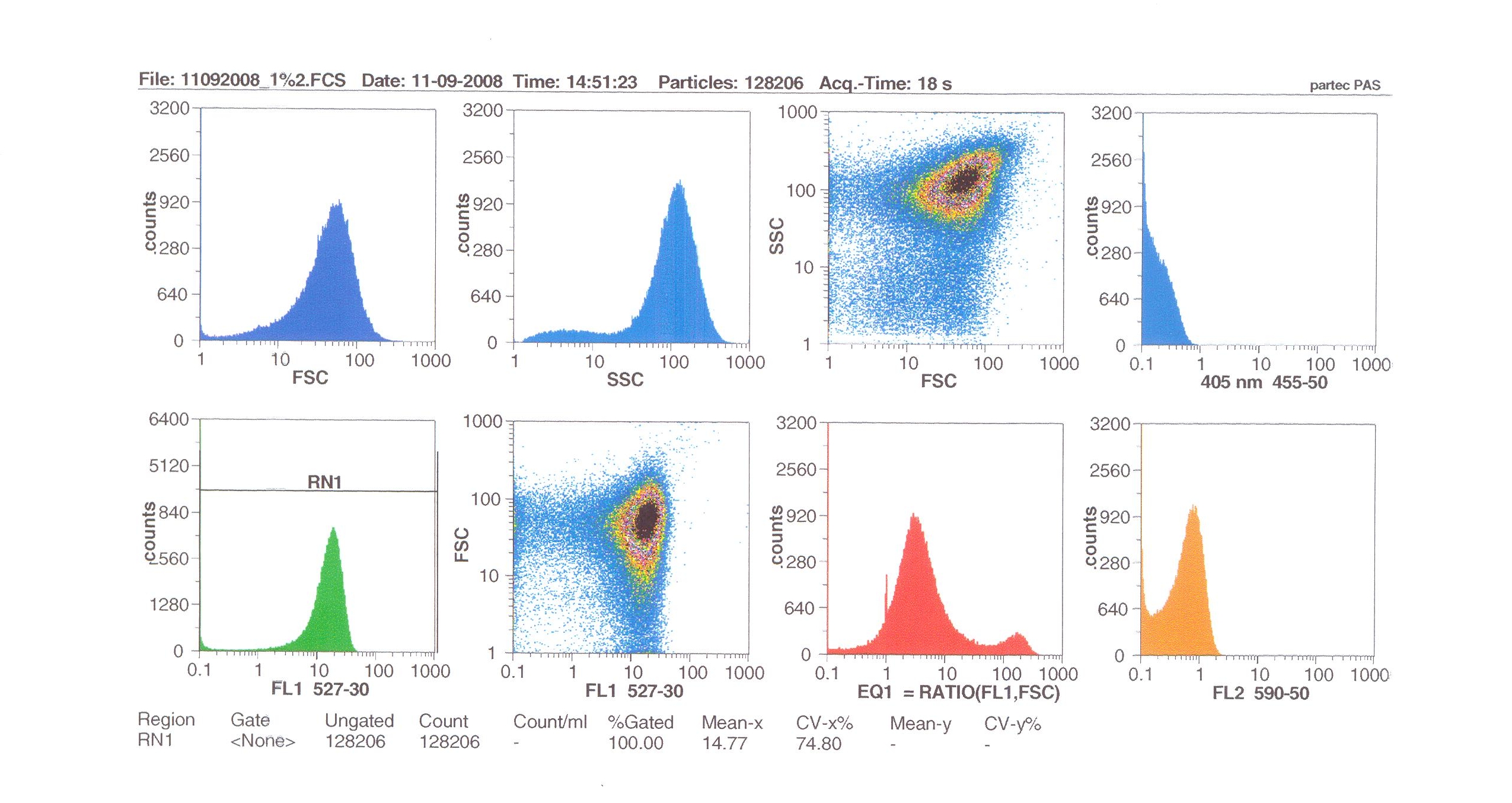
| + | '''Induction by 1% v/v subtilin:''' mean fluoresence = 14.77 | ||
| + | |||
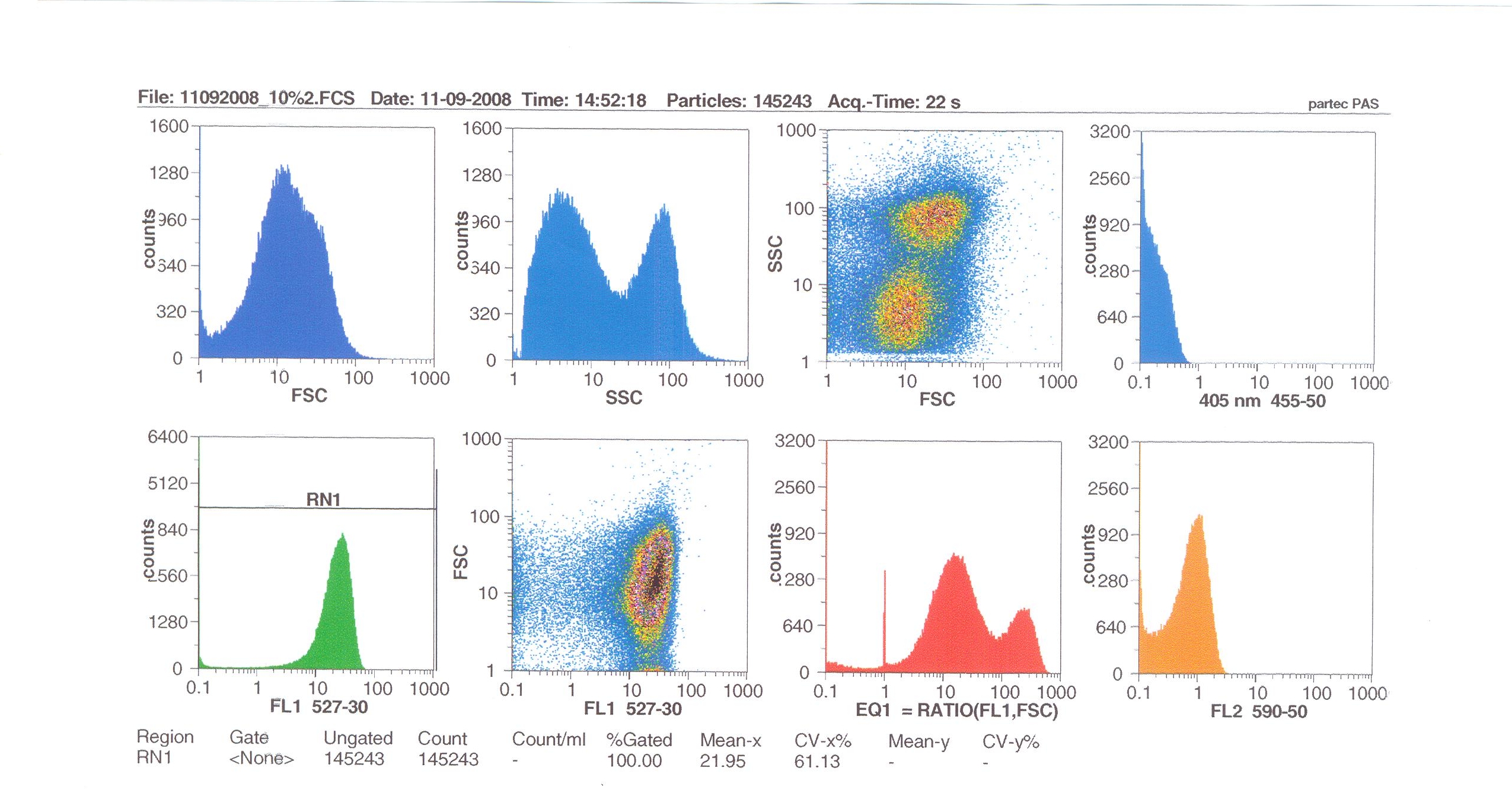
| + | '''Induction by 10% v/v subtilin:''' the mean fluoresence = 21.95 | ||
| + | |||
| + | These results show that the higher the concentration of subtilin, the more GFP is expressed. | ||
| + | |||
| + | At 10% induction, the subtilin began to cause some of the cells to lyse - this can be seen by the two distinct populations of cells - one being the cells which have lysed, and the other being the remainder of intact cells. Since subtilin is an antibiotic, this was a good indication that the subtilin we had collected that day was of good quality. | ||
| + | |||
| + | Overall, the microscopy and flow cytometry suggest that our biobrick is functional, but requires high levels of subtilin in order to be activated. The days that we performed microscopy for instance, we did not observe killing of the cells after 10% addition of subtilin, indicating that our subtilin for that day was at a low concentration. Subsequently, we did not observe a strong GFP induction. However, when we did observe killing (indicative of high subtilin concentrations), we did observe induction of GFP (see flow cytometry). So in principle the system works, and we have engineered the lab strain of ''B. subtilis'' 168 to respond to the external addition of subtilin, but more work should be done to further optimise the system. For instance, it was shown that the spaS promoter fused to gfp gives high yield of protein production when present on a multicopy plasmid, after the addition of subtilin (Bongers et al, AEM). Also it may be possible to introduce a positive feedback effect by introducing a ''spaS'' box and associated promoter upstream of ''spaRK'' in addition to the normal promoter. | ||
| + | |||
| + | <gallery> | ||
| + | Image:Montage.jpg|Microscopy results of gfp upon 1% induction by subtilin | ||
| + | Image:Montage_pc.jpg|Microscopy results of mCherry upon 1% induction by subtilin | ||
| + | Image:Flow0%.jpg|Flow cytometry results for 0% induction by subtilin. | ||
| + | Image:Flow1%.jpg|Flow cytometry results for 1% induction by subtilin. | ||
| + | Image:Flow10%.jpg|Flow cytometry results for 10% induction by subtilin. | ||
| + | </gallery> | ||
| + | |||
| + | |||
| + | |||
| + | <html><a name="protocols"></html> | ||
| + | ===Protocols=== | ||
| + | |||
| + | Listed below are the lab protocols that were developed or used for the Newcastle University iGEM 2008 wet lab component. Please click on any item for detailed instructions. | ||
| - | + | * [[Agarose Gel Electrophoresis]] | |
| + | * [[Making Agar Plates]] | ||
| + | * [[Isolating Plasmid from Cells (Miniprep)]] | ||
| + | * [[Restricting Plasmids (Double Restriction)]] | ||
| + | * [[Purifying DNA from an enzymatic reaction]] | ||
| + | * [[Cutting a Specific Band from Agarose Gel]] | ||
| + | * [[Purifying DNA from Gel Slices]] | ||
| + | * [[Ligating DNA]] | ||
| + | * [[Transforming into DH5α or TOP10 E. coli|Transforming into DH5α or TOP10 ''E. coli'']] | ||
| + | * [[Making Frozen Glycerol Cell Stocks From Overnight Cultures]] | ||
| + | * [[Making Overnight Cultures from Frozen Glycerol Cell Stocks]] | ||
| + | * [[Making Overnight Cultures from Agar Colonies]] | ||
| + | * [[Transforming into Bacillus subtillis|Transforming into ''Bacillus subtilis'']] | ||
| + | * [[Inducing Bacillus subtilis with Subtilin|Inducing ''Bacillus subtilis'' with Subtilin]] | ||
| + | * [[Preparing Bacillus subtilis for Microscopy|Preparing ''Bacillus subtilis'' for Microscopy]] | ||
| - | |||
| - | |||
| - | + | <html><a name="journal"></html> | |
| + | == Wet Lab Journal == | ||
| - | + | This contains the day-to-day progress of our wet lab team. Click on a day below to find more details of the work above. | |
| + | {|cellpadding="10" | ||
| + | |{{#calendar: title=Newcastle University Wetlab |year=2008 | month=08 }} | ||
| + | |{{#calendar: title=Newcastle University Wetlab |year=2008 | month=09 }} | ||
| + | |} | ||
| - | + | </div><!--main col --> | |
| - | </div><!-- | + | |
| - | + | ||
Latest revision as of 01:53, 30 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Wet Lab
Introduction
The overall design was too complicated for the time and resources available so we devised a proof of concept Brick which represented an input node from our full design. This Brick demonstrates that it is possible to transfer two-component peptide sensing from one strain into another and have it function as it did in the first strain. We took the subtilin two component system from Bacillus subtilis ATCC6633 and integrated it into the chromosome of B. subtilis 168, the typical laboratory B. subtilis strain at the amyE locus.
The wet lab work involved:
- Cloning the synthesized Brick from pUC57 in E. coli into our Bacillus integration vectors (with GFP and mCherry respectively downstream of the Brick)
- Validation of the integration vector
- Transformation and integration into the chromosome of B. subtilis 168 at the amyE locus
- Validation of chromosomal integration
- Characterisation of the integrated parts and their response to subtilin.
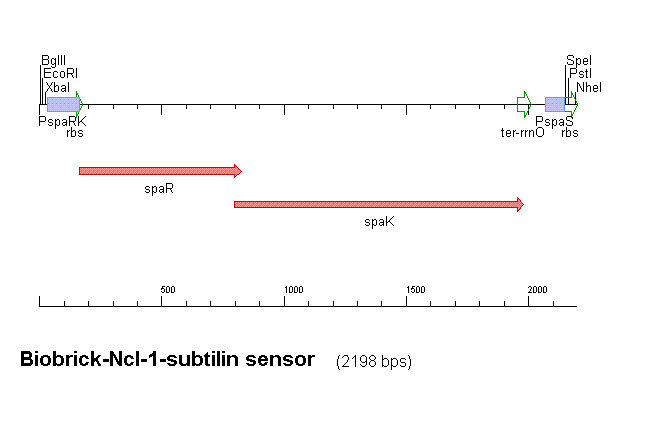
Our subtilin sensor, part [http://partsregistry.org/Part:BBa_K104001 BBa_k104001]
Figure 1 shows the construct which contains:
- spaRK promotor
- spaR (subtilin peptide antibiotic Regulation) - the 220 amino acid product of this gene usually regulates the downstream production of subtilin antibiotic. It has an N-terminal domain that can be phosphorylated and a C-terminal domian that has DNA binding properties [http://aem.asm.org/cgi/reprint/59/1/296.pdf]
- spaK (subtilin peptide antibiotic Kinase) - this gene codes for a 325 amino acid histadine kinase peptide that phosphorylates the N-terminus of spaR [http://aem.asm.org/cgi/reprint/59/1/296.pdf]. This activates the DNA binding ability of the C-terminus of spaR, which in turn initiates transcription of the downstream gene. In the case of our construct, this gene is gfp.
- rrnO transcriptional terminator
- spaS promotor - a strong promotor inducible by upstream activation of spaRK. It can be placed in front any gene to regulate its activity.
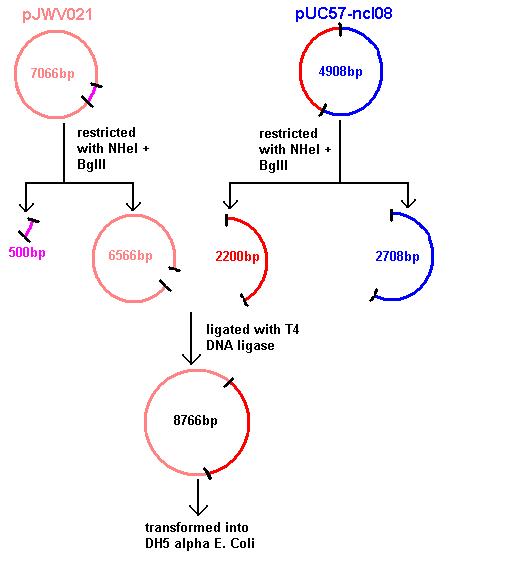
Aim 1: clone the spaRK system from pUC57 into pJWV021 and transform into DH5 alpha competent E. coli
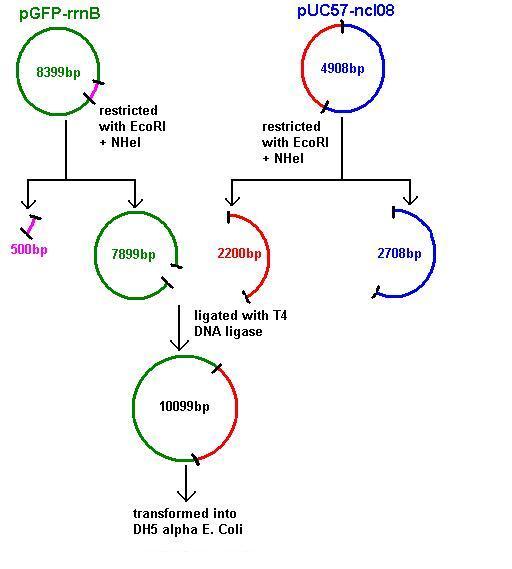
Aim 2: clone the spaRK system from pUC57 into pGFP-rrnB and transform into DH5 alpha competent E. coli
- The 2.2kb fragment (ncl08) contains the spaRK biobrick system. When properly cloned, our biobrick will replace the rrnB promoter in front of gfp present in plasmid pGFP-rrnB. Thus, gfp will now be under control of the subtilin inducible spaS promoter. This means that when SpaR is activated by SpaK (by sensing subtilin) its positive regulatory effect on PspaS will activate gfp.
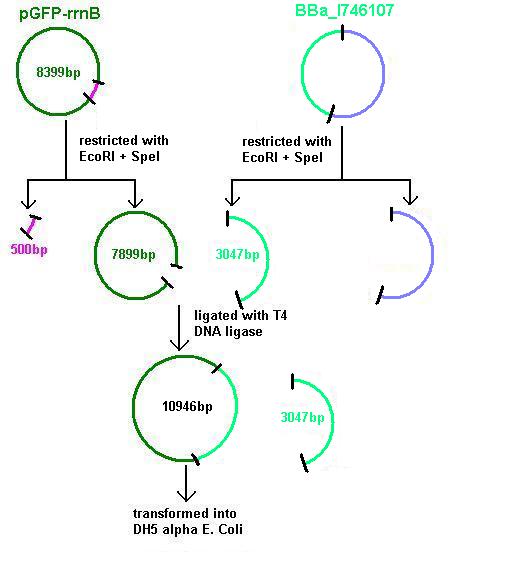
Aim 3: clone the agr (Cambridge biobrick for sensing S. aureus) system from the biobrick plasmid using EcoRI and SpeI sites into the corresponding sites of pGFP-rrnB. This will replace the PrrnB-gfp on the bacillus vector with the agr biobrick. The ligation was transformed into DH5 alpha competent E. coli
- BBa_I746107 contains a partial agr operon, which includes agrC and A but has agrB and D deleted. This allows coding for the receptor (C/A) but not for production of the quorum peptides themselves (B/D). The inducible promoter drives expression of gfp.
Subcloning
The pUC57 was restriction-digested and the fragments displayed on a gel to ensure that the correct products have been produced.
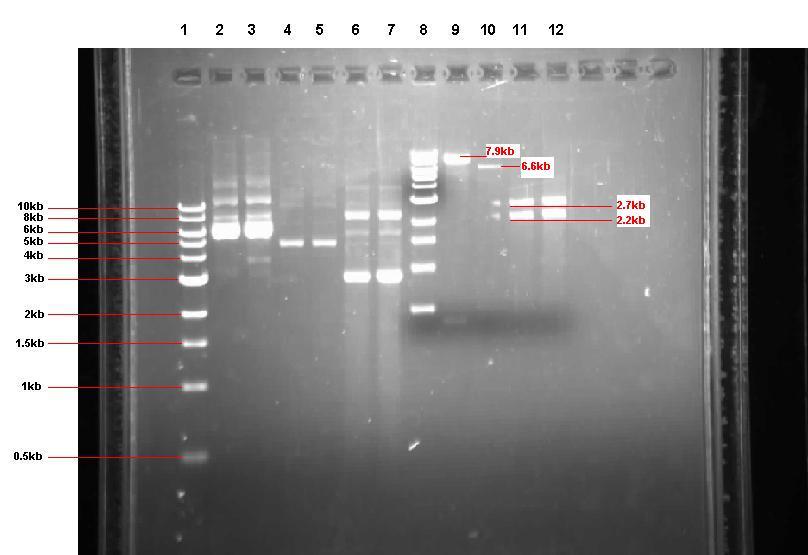 Figure 2: Gel from 2 Sept showing our 3 unrestricted plasmids, along with the 4 restrictions
Figure 2: Gel from 2 Sept showing our 3 unrestricted plasmids, along with the 4 restrictions
- Lane 1: 1kb ladder
- Lane 2: Unrestricted pGFP-rrnB sample 1 (our bacillus integration vector that carries gfp between 2 amyE flanking regions)
- Lane 3: Unrestricted pGFP-rrnB sample 2
- Lane 4: Unrestricted pJWV021 sample 1 (our bacillus integration vector that carries mCherry between 2 sacA flanking regions)
- Lane 5: Unrestricted pJWV021 sample 2
- Lane 6: Unrestricted pUC57-ncl08 sample 1 (our synthetic biobrick construct; GenScript)
- Lane 7: Unrestricted pUC57-ncl08 sample 2
- Lane 8: 1kb ladder
- Lane 9: pGFP-rrnB restricted with EcoR1 and Nhe1
- Lane 10: pJWV021 restricted with BglII and Nhe1
- Lane 11: pUC57-ncl08 restricted with EcoR1 and Nhe1
- Lane 12: pUC57-ncl08 restricted with BglII and Nhe1
Not shown here is the agr biobrick digestion.
Validation of the integration vector
Digested pGFP-rrnB/pJWV021 were mixed with digested pUC57-ncl08 or agr biobrick, ligated and transformed to E.coli and the bacteria overnight were grown on selective media (the pGFP ligation on plates with spectinomycin (50microG/ml) and the pJWV021 ligation on kanamycin (10 microG/ml)). Transformants were grown up in LB with antibiotics, plasmid was isolated and digested to check for successful subcloning (in the GFP and mCherry vectors, GFP results shown below).
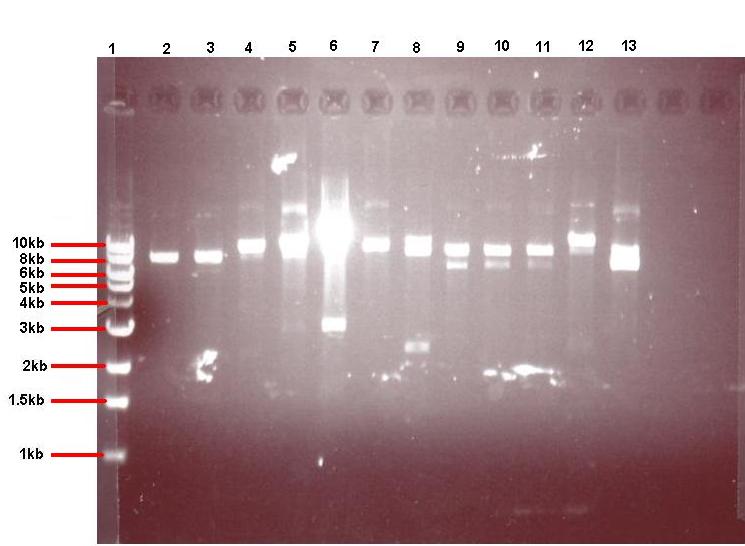 Figure 4: Gel showing restricted pGFPrrnB-ncl08 taken from 12 different colonies
Figure 4: Gel showing restricted pGFPrrnB-ncl08 taken from 12 different colonies
- Lane 1: 1kb ladder
- Lane 2: pGFPrrnB-ncl08 colony 1 (white colony, 25μl plate)
- Lane 3: pGFPrrnB-ncl08 colony 2 (white colony, 25μl plate)
- Lane 4: pGFPrrnB-ncl08 colony 3 (white colony, 25μl plate)
- Lane 5: pGFPrrnB-ncl08 colony 4 (white colony, 25μl plate)
- Lane 6: pGFPrrnB-ncl08 colony 5 (white colony, 25μl plate)
- Lane 7: pGFPrrnB-ncl08 colony 6 (white colony, 25μl plate)
- Lane 8: pGFPrrnB-ncl08 colony 7 (white colony, 25μl plate)
- Lane 9: pGFPrrnB-ncl08 colony 8 (white colony, 25μl plate)
- Lane 10: pGFPrrnB-ncl08 colony 9 (white colony, 25μl plate)
- Lane 11: pGFPrrnB-ncl08 colony 10 (white colony, 250μl plate)
- Lane 12: pGFPrrnB-ncl08 colony 11 (white colony, 250μl plate)
- Lane 13: pGFPrrnB-ncl08 colony 12 (green colony, 25μl plate)
This demonstrated that we had succesfully subcloned our ncl08 biobrick from pUC57-ncl08 to pGFP. Not shown here are the restriction checks for pJWV021-ncl08 and pGFP-agr (but we have correct clones for them as well!).
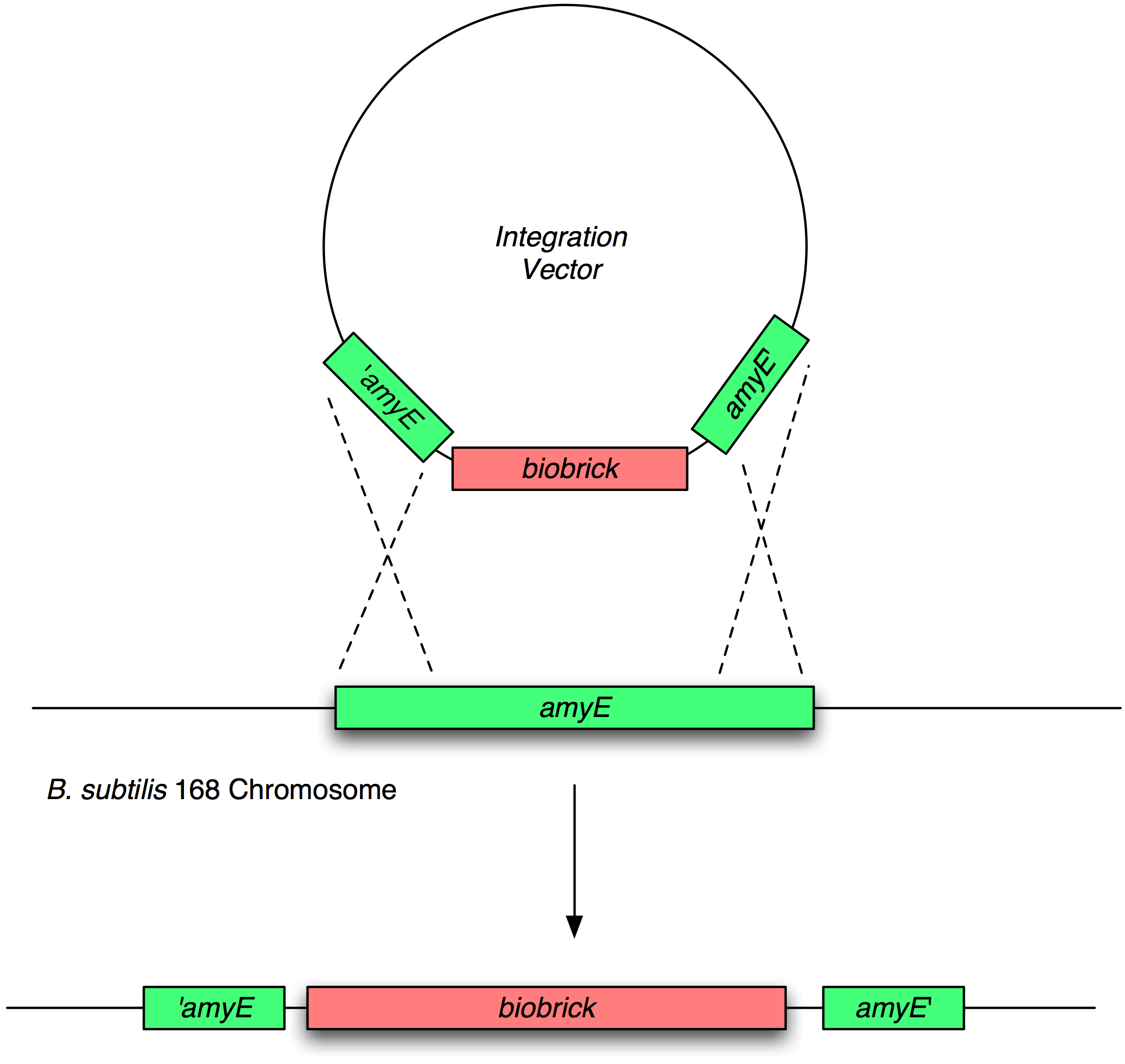
Transformation and integration into the B. subtilis chromosome
Validation of chromosomal integration
Next, we transformed the correct plasmids to (self made) competent Bacillus, as described in the protocol section. For plasmid pGFP-ncl08 and pGFP-agr, bacillus was plated on chloramphenicl (5 microG/ml) and for plasmid pJWV021-ncl08, bacillus were plated on kanamycin (5 microG/ml). Next day, Bacillus transformants were subseqently streaked to single colonies on selective agar plates. To check for correct integration of the pGFP-ncl08 and pGFP-agr plasmids at the amyE locus, single colonies were streaked on nutrient agar plates containing 1% starch. If a double crossover event occured, cells are not able to produce AmyE and will not degrade starch, thus no 'halo' around the colony. 2 individual colonies without an halo were grown and stocked and used for later analyses. To check correct integration of the pJWV021-ncl08 plasmid in the Bacillus chromosome, single colonies were streaked on minimal medium plates containing either glucose or sucrose as the sole carbon source. If the plasmid integrated correctly at the sacA locus via a double crossover, cells cannot utilise sucrose anymore and will not grow on the minimal plate containing sucrose, but will grow on the glucose plate. Again, 2 correct transformants were selected that were unable to proliferate on sucrose plates and stocked in the -80C freezer for future work.
Characterisation of the integrated parts and their response to subtilin
Results
To analyse the results of the wet lab transformations of the inserts into B. subtilis, we used two methods: microscopy and flow cytometry.
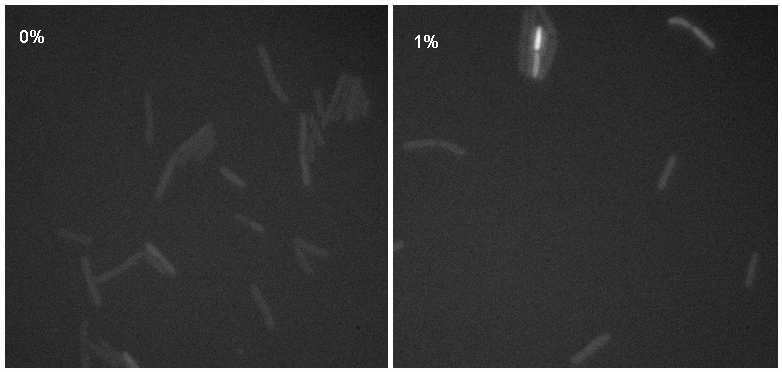
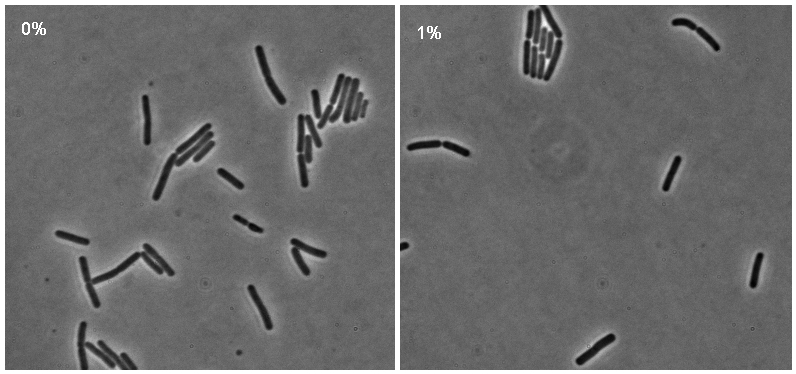
Microscopy
Microscopy work from 08.09.08 showed a difference in the level of flourescence of the iGEMgfp fluorescent cells (higher in 10% subtilin-induced cells compared to 0% subtilin-induced cells). However, there was little difference in the number of cells that fluoresced between the two cultures.
There was no difference in the number of fluorecent cells or the level of fluorescence between the 10% subtilin-induced and the 0% subtilin-induced iGEMcherry cells.
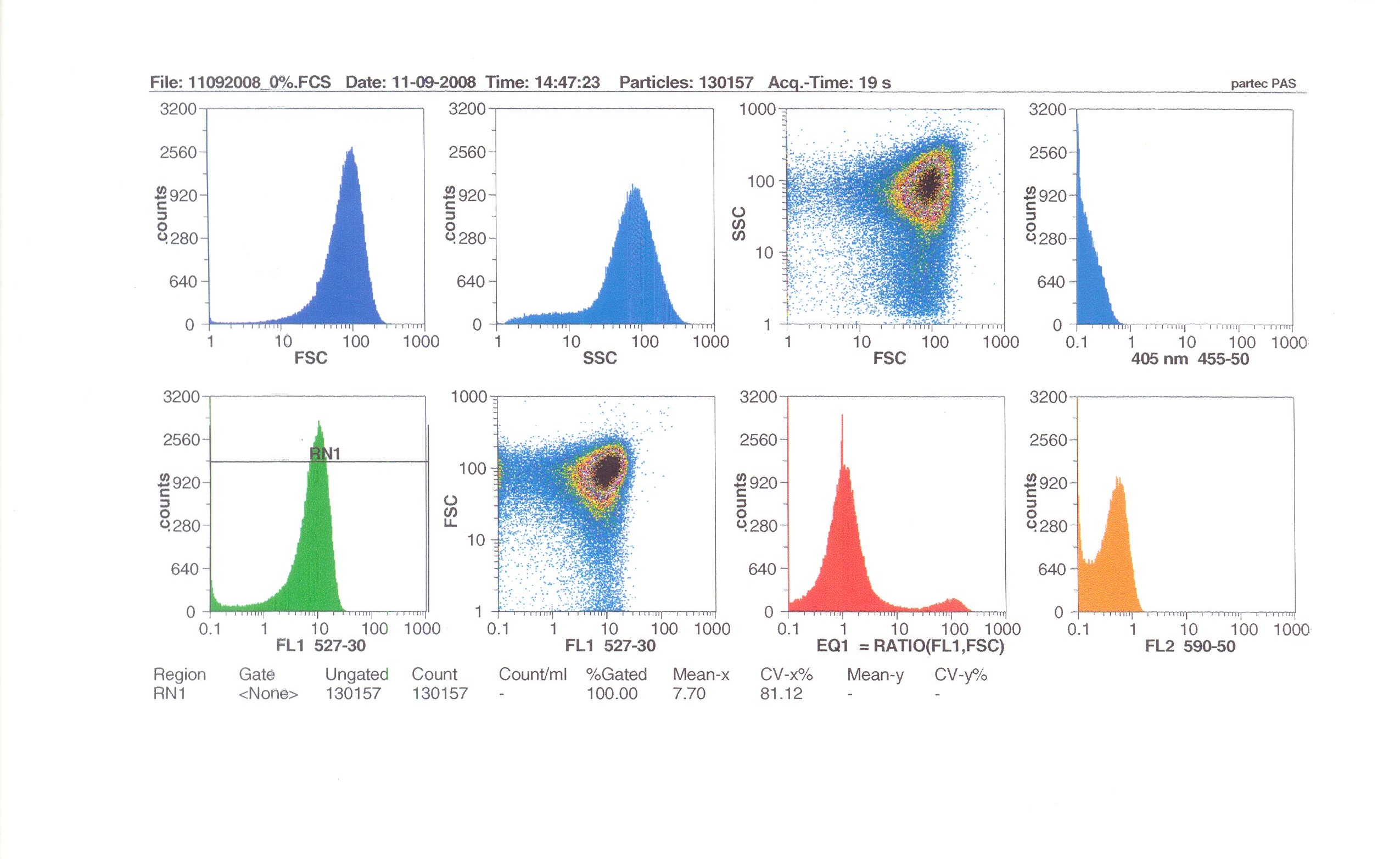
Flow cytometry
Flow cytometry measures cell fluorescence and light scattering, cell-by-cell, allowing us to quantify our results and present them in graphical form. A sample of our cells engineered B. subtilis cells were injected into the machine, which hydro-dynamically focusses the fluid. Lasers of a particular wavelength are directed onto the stream of fluid, and each particle which passes through the light beam will cause the laser to scatter in a different way. Cells which absorb and then re-emit the light, result in fluorescence (a higher energy state).
The detectors in the machine measure the scattering of light and any fluoresence which occurs, and the data is displayed as histograms and dot plots.
Induction by 0% v/v subtilin (i.e in the absence of subtilin): mean fluoresence = 7.70
Induction by 1% v/v subtilin: mean fluoresence = 14.77
Induction by 10% v/v subtilin: the mean fluoresence = 21.95
These results show that the higher the concentration of subtilin, the more GFP is expressed.
At 10% induction, the subtilin began to cause some of the cells to lyse - this can be seen by the two distinct populations of cells - one being the cells which have lysed, and the other being the remainder of intact cells. Since subtilin is an antibiotic, this was a good indication that the subtilin we had collected that day was of good quality.
Overall, the microscopy and flow cytometry suggest that our biobrick is functional, but requires high levels of subtilin in order to be activated. The days that we performed microscopy for instance, we did not observe killing of the cells after 10% addition of subtilin, indicating that our subtilin for that day was at a low concentration. Subsequently, we did not observe a strong GFP induction. However, when we did observe killing (indicative of high subtilin concentrations), we did observe induction of GFP (see flow cytometry). So in principle the system works, and we have engineered the lab strain of B. subtilis 168 to respond to the external addition of subtilin, but more work should be done to further optimise the system. For instance, it was shown that the spaS promoter fused to gfp gives high yield of protein production when present on a multicopy plasmid, after the addition of subtilin (Bongers et al, AEM). Also it may be possible to introduce a positive feedback effect by introducing a spaS box and associated promoter upstream of spaRK in addition to the normal promoter.
Protocols
Listed below are the lab protocols that were developed or used for the Newcastle University iGEM 2008 wet lab component. Please click on any item for detailed instructions.
- Agarose Gel Electrophoresis
- Making Agar Plates
- Isolating Plasmid from Cells (Miniprep)
- Restricting Plasmids (Double Restriction)
- Purifying DNA from an enzymatic reaction
- Cutting a Specific Band from Agarose Gel
- Purifying DNA from Gel Slices
- Ligating DNA
- Transforming into DH5α or TOP10 E. coli
- Making Frozen Glycerol Cell Stocks From Overnight Cultures
- Making Overnight Cultures from Frozen Glycerol Cell Stocks
- Making Overnight Cultures from Agar Colonies
- Transforming into Bacillus subtilis
- Inducing Bacillus subtilis with Subtilin
- Preparing Bacillus subtilis for Microscopy
Wet Lab Journal
This contains the day-to-day progress of our wet lab team. Click on a day below to find more details of the work above.
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"