Team:Hawaii/Notebook/2008-07-29
From 2008.igem.org
(Difference between revisions)
(→Plasmid prep) |
|||
| (6 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
===Construction of BioBricks=== | ===Construction of BioBricks=== | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
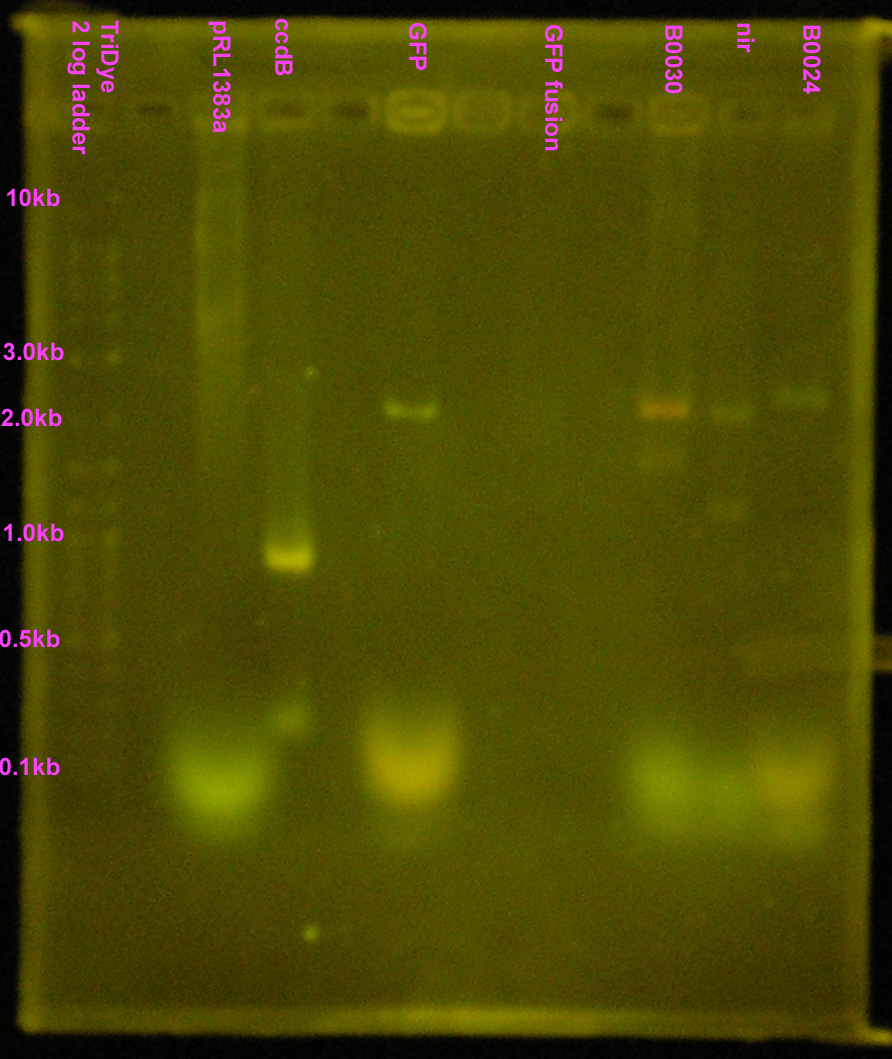
| - | + | [[Image:072908REdigests.jpg|right|200px|thumb|EtBr stained 1.2% agarose gel. Thirty microliters of each RE reaction and 10 microliters of ccdB were loaded into wells. Gel ran at 60V for 2 hours.]] | |
| - | :* Purified RE digests on EtBr stained | + | :* Purified RE digests on EtBr stained 1.2% agarose gel |
::* Extracted bands for B0030 and B0024 | ::* Extracted bands for B0030 and B0024 | ||
::* Other lanes FAILED! | ::* Other lanes FAILED! | ||
| + | ::* SYBR Safe doesn't show up well, even under UV (arg...) | ||
===Direct Band Extraction Method=== | ===Direct Band Extraction Method=== | ||
| + | :<strong>Norman</strong> | ||
:* make two comb gels | :* make two comb gels | ||
:* run test UPA band along with ladder | :* run test UPA band along with ladder | ||
| Line 34: | Line 36: | ||
:*Amplified with Green Taq: rep region | :*Amplified with Green Taq: rep region | ||
| - | :*On [[Team:Hawaii/Notebook/2008- | + | :*On [[Team:Hawaii/Notebook/2008-07-30|7-30]] I ran a gel. |
===Media=== | ===Media=== | ||
| - | + | :<strong>Margaret</strong> | |
:*started to make LB amp100 plates, but autoclave room is closed. Remember to do it tomorrow. | :*started to make LB amp100 plates, but autoclave room is closed. Remember to do it tomorrow. | ||
| - | + | ===Plasmid prep=== | |
| - | + | :<strong>Krystle</strong> | |
| + | :* Innoculated LB+amp with pRL1383a harboring E. coli (from cryostock) | ||
| + | ===PCR=== | ||
| + | :<strong>Krystle</strong> | ||
| + | :* PCR reactions to isolate GFP, GFPf, nir from plasmid | ||
= Discussion = | = Discussion = | ||
Latest revision as of 09:40, 31 July 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Gel purification of ccdB PCR product
- Grace
RE digests
- Grace
- Digested pRL1383a (from last night) with BamHI
- Digested B0030 and B0024 (from last night) with EcoRI
- Sequentially digested gel purified ccdB with HindIII and BamHI
Construction of BioBricks
- Grace
- Purified RE digests on EtBr stained 1.2% agarose gel
- Extracted bands for B0030 and B0024
- Other lanes FAILED!
- SYBR Safe doesn't show up well, even under UV (arg...)
Direct Band Extraction Method
- Norman
- make two comb gels
- run test UPA band along with ladder
- attempt to pipette out DNA as it runs into the second well
- no more gel cutting???
PCR amplification of pRL1383a parts
- Margaret
- Made a reaction buffer containing {38.5ul, 20ul accusure, 10ul dNTPs, & 5ul 5XBuffer}. This should be good for 20 50ul reactions.
- Amplified with Accusure: aadA ([http://partsregistry.org/Part:BBa_J23012 BBa_J23012]), aadaA (pRL1383a), rep region, oriV
- Amplified with Green Taq: rep region
- On 7-30 I ran a gel.
Media
- Margaret
- started to make LB amp100 plates, but autoclave room is closed. Remember to do it tomorrow.
Plasmid prep
- Krystle
- Innoculated LB+amp with pRL1383a harboring E. coli (from cryostock)
PCR
- Krystle
- PCR reactions to isolate GFP, GFPf, nir from plasmid
Discussion
SYBR Safe
- Blue light still doesn't illuminate the bands well. No bands (not even the ladder) are visible. Resolution *much* better with short wave UV. EtBr better bet for now?
Restriction enzymes
- HindIII and BamHI in the lab -20C freezer (in box labeled "TKW Restriction Enzymes") fail. They're really old and don't appear to cut well anymore. Digestion of pRL1383a resulted in a long smear.
Quote of the Day
LOL! http://www.youtube.com/watch?v=J0s0Y3-BCaw
 "
"