Team:Hawaii/Notebook/2008-08- 4
From 2008.igem.org
m |
(→PCR) |
||
| (3 intermediate revisions not shown) | |||
| Line 28: | Line 28: | ||
<strong> Results </strong> | <strong> Results </strong> | ||
| - | Lane 2: PCR verification of I14032: There are two bands. The top | + | <strong>Lane 2:</strong> PCR verification of I14032: There are two bands. The top |
band does not correspond to Plac, while the bottom band is just above | band does not correspond to Plac, while the bottom band is just above | ||
0.4kb, about 50bp more than would be expected. | 0.4kb, about 50bp more than would be expected. | ||
| - | Lane 3: PCR verification of OriT: The band is just above 0.4kb, and the | + | <strong>Lane 3:</strong> PCR verification of OriT: The band is just above 0.4kb, and the |
verification part is 0.415 kb. | verification part is 0.415 kb. | ||
| - | Lane 4: Amplification of oriV from pRL1383a:The band is at 4.5kb, as | + | <strong>Lane 4:</strong> Amplification of oriV from pRL1383a:The band is at 4.5kb, as |
expected. | expected. | ||
| - | Lane 5&8: pSB1A7 was used as a (+) control: While the same size was amplified | + | <strong>Lane 5&8:</strong> pSB1A7 was used as a (+) control: While the same size was amplified |
in both positive control reactions, I would expect a band at 1265bp, but | in both positive control reactions, I would expect a band at 1265bp, but | ||
the amplicon is just below 0.9kb. | the amplicon is just below 0.9kb. | ||
| - | Lane 6&9: VF2 and VR were used to amplify water. As expected, nothing was amplified. | + | <strong>Lane 6&9:</strong> VF2 and VR were used to amplify water. As expected, nothing was amplified. |
| - | Lane 7:The rep region was amplified correctly. I expected a band at 3.352kb and the band | + | <strong>Lane 7:</strong>The rep region was amplified correctly. I expected a band at 3.352kb and the band |
occurred in this region. | occurred in this region. | ||
<strong> Discussion </strong> | <strong> Discussion </strong> | ||
| - | :* I did some trouble shooting on this PCR with advice from: [[http://www.med.yale.edu/genetics/ward/tavi/Trblesht.html]]. | + | |
| + | :*I did some trouble shooting on this PCR with advice from: [[http://www.med.yale.edu/genetics/ward/tavi/Trblesht.html]]. | ||
:*In most cases, I just got as close to the recommended annealing temperature and extension time in order to decrease the amount of unspecific products. | :*In most cases, I just got as close to the recommended annealing temperature and extension time in order to decrease the amount of unspecific products. | ||
:*I only ran 30 cycles as opposed to 35 in the past. I think this has made the efficiency of my taq more consistent, especially with the longer products. | :*I only ran 30 cycles as opposed to 35 in the past. I think this has made the efficiency of my taq more consistent, especially with the longer products. | ||
| Line 53: | Line 54: | ||
:* pSMC121 (we were running out of template) | :* pSMC121 (we were running out of template) | ||
:* re-streaked transformants on 8-5. There was a lawn of transformants. | :* re-streaked transformants on 8-5. There was a lawn of transformants. | ||
| - | |||
===Re-streak Library=== | ===Re-streak Library=== | ||
| Line 61: | Line 61: | ||
===Construction of p+r, g+t=== | ===Construction of p+r, g+t=== | ||
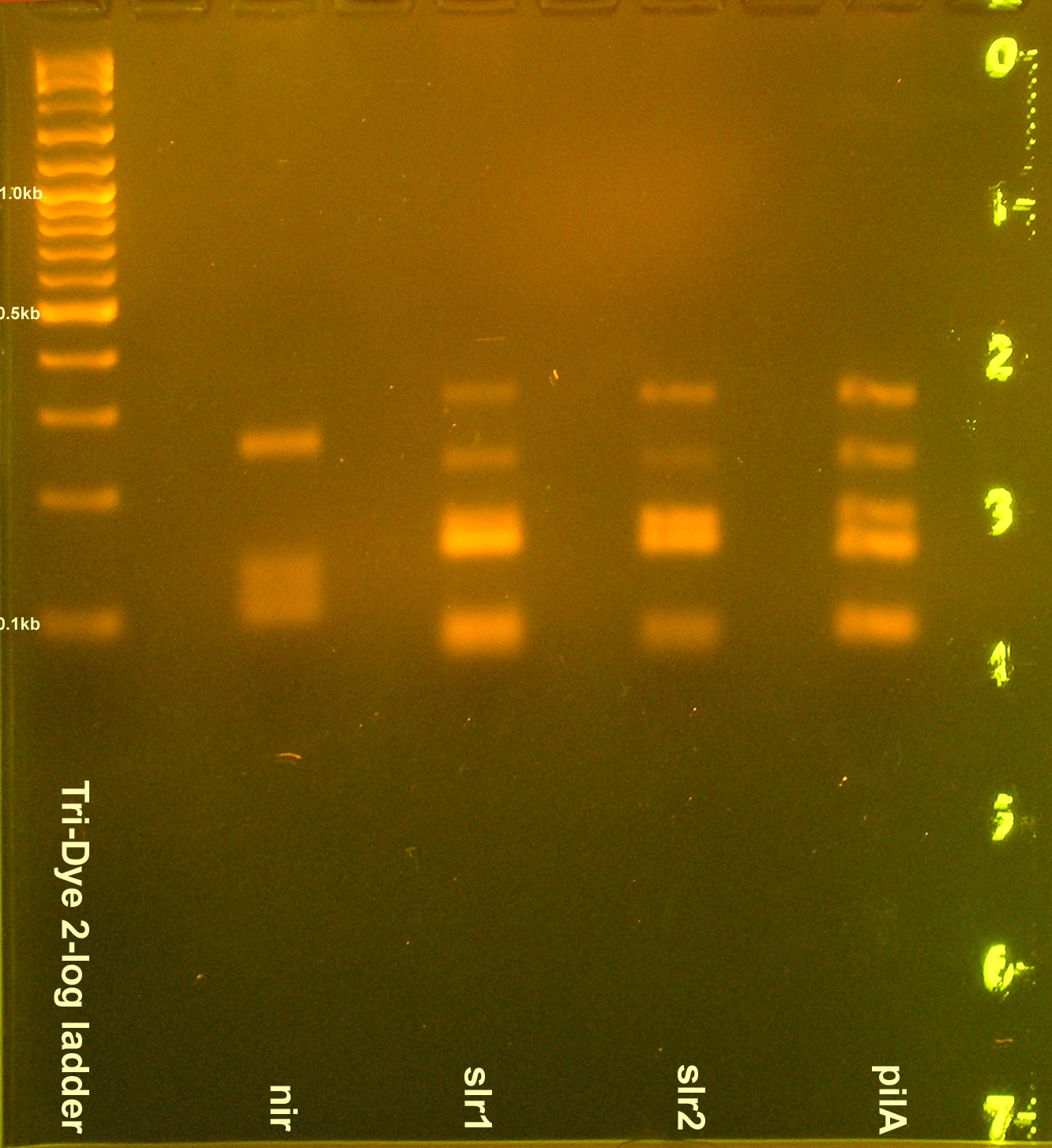
| + | [[Image:080408PCRREnirsig.jpg|right|thumb|200px|EtBr stained 3% agarose gel ran at 60V for 100 min. Twenty microliters of the restriction digests were loaded into each well.]] | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
| Line 82: | Line 83: | ||
=== PCR=== | === PCR=== | ||
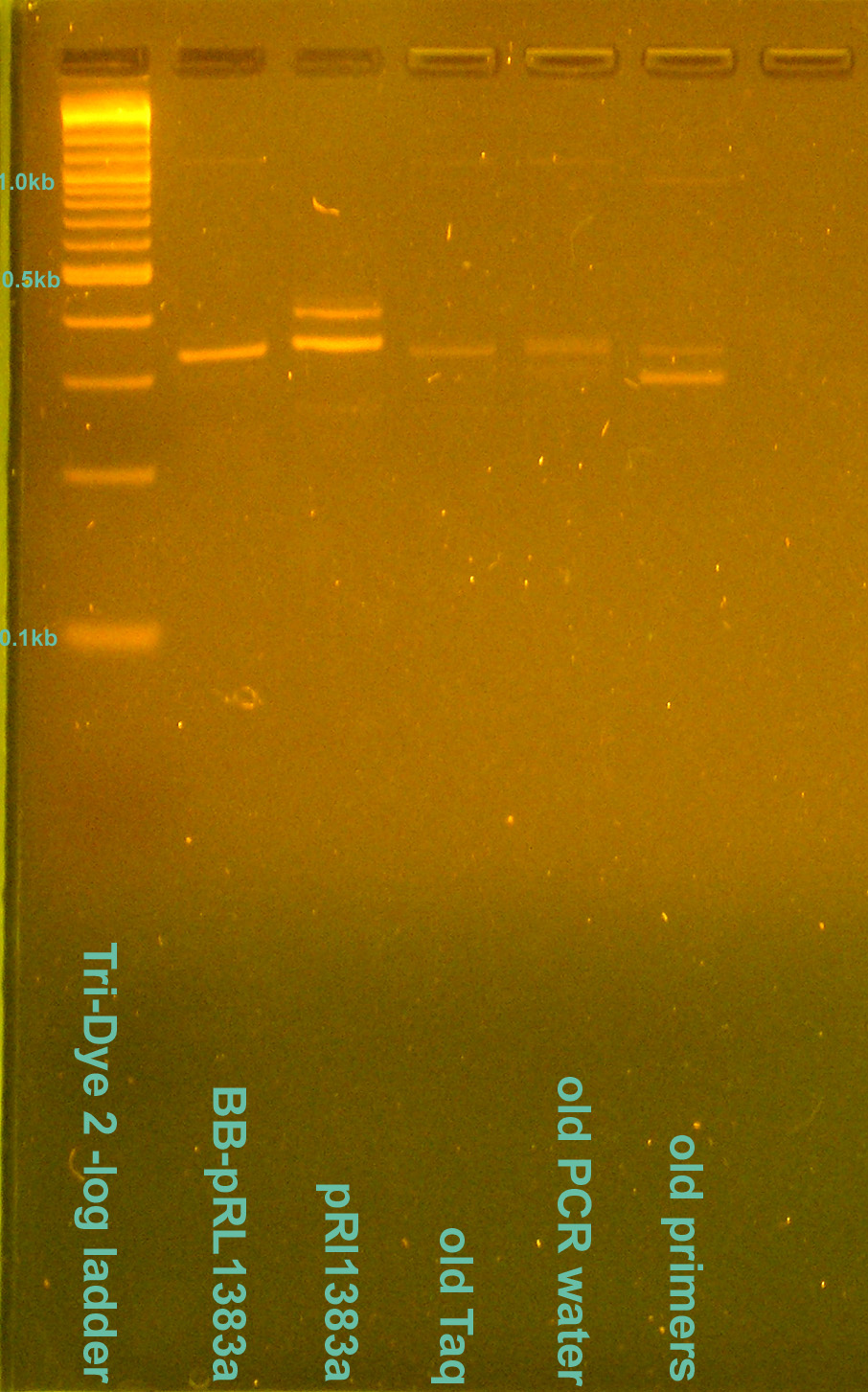
| + | [[Image:080408PCRcontamination.jpg|right|thumb|200px|EtBr stained 2.5% agarose gel ran at 95V for 1 hour. Ten microliters of the PCR reactions were loaded into each well.]] | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
| - | |||
:* Redid Saturday's PCR of BB-pRl1383a and pRL1383a alongside test of PCR contamination | :* Redid Saturday's PCR of BB-pRl1383a and pRL1383a alongside test of PCR contamination | ||
| Line 89: | Line 90: | ||
:::* Gel doesn't match that from Saturday | :::* Gel doesn't match that from Saturday | ||
:* Redid PCR again to verify Saturday/today's PCR | :* Redid PCR again to verify Saturday/today's PCR | ||
| - | |||
| - | |||
= Discussion = | = Discussion = | ||
Latest revision as of 08:03, 7 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Plasmid Prep
- Margaret
- I14032, oriT
Quantification of Parts with Gel
- Margaret
- I14032, oriT, L51020, pSB1A7, B015, B0034
PCR
- Margaret
- Rep, oriV, verification of I14032, oriT(just to make sure)
Materials & Methods
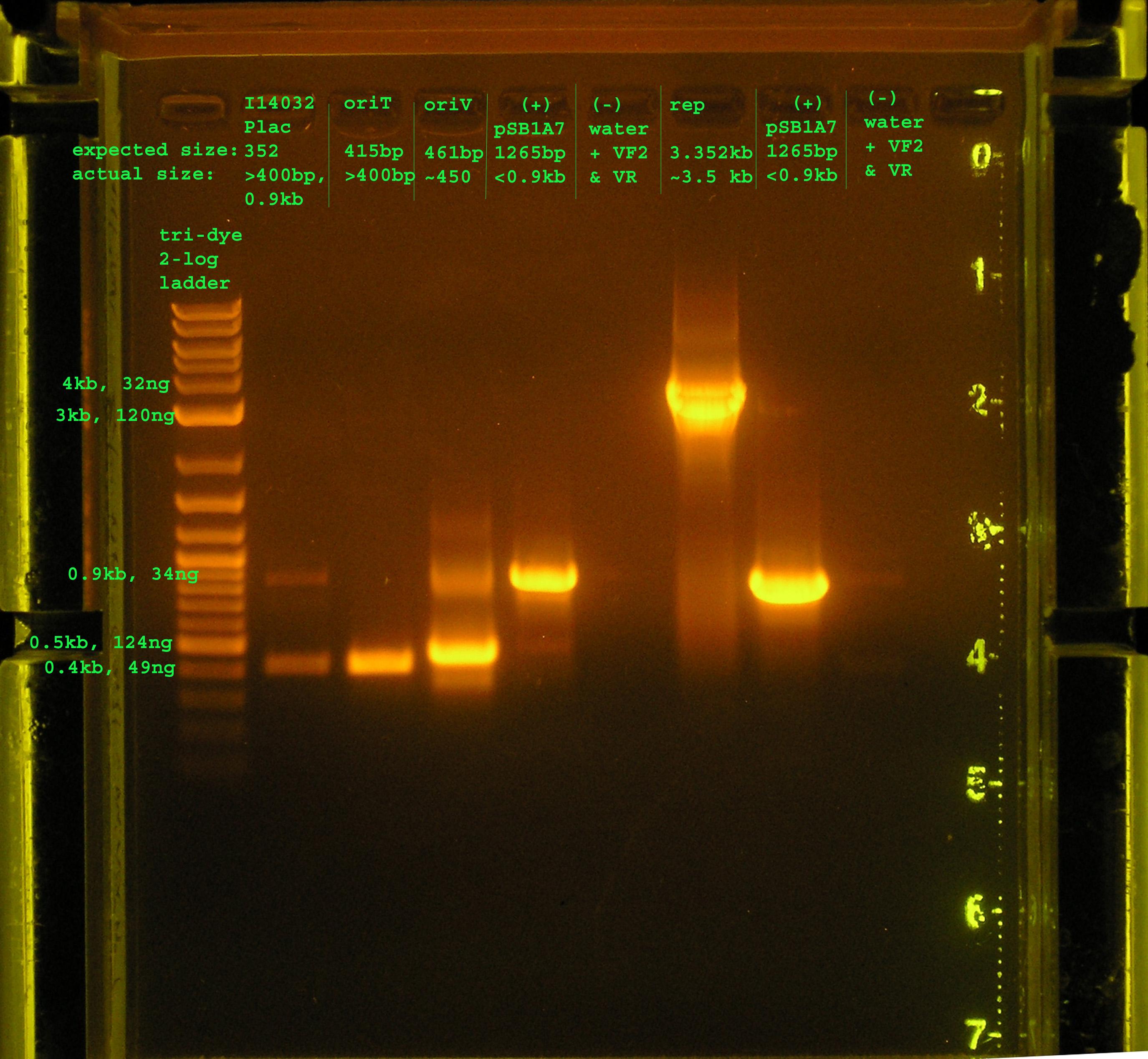
- Reaction Conditions: 3.5ul water, 0.5ul template, 1.0ul f&r primer, 5ul econotaq
- Running conditions (I14032, oriT): 94°C hold, 94°C 2', (95°C 30", 62°C 30", 72°C 30") x 30cycles, 72°C 10', 4°C hold
- Running conditions (oriV + controls): 94°C hold, 94°C 2', (95°C 30", 51.3°C 30", 72°C 30") x 30cycles, 72°C 10', 4°C hold
- Running conditions (rep + controls): 94°C hold, 94°C 2', (95°C 30", 53.9°C 30", 72°C 3'20") x 20 cycles, (95°C 30", 53.9°C 30", 72°C 3'30") x 10 cycles72°C 10', 4°C hold
Results
Lane 2: PCR verification of I14032: There are two bands. The top band does not correspond to Plac, while the bottom band is just above 0.4kb, about 50bp more than would be expected. Lane 3: PCR verification of OriT: The band is just above 0.4kb, and the verification part is 0.415 kb. Lane 4: Amplification of oriV from pRL1383a:The band is at 4.5kb, as expected. Lane 5&8: pSB1A7 was used as a (+) control: While the same size was amplified in both positive control reactions, I would expect a band at 1265bp, but the amplicon is just below 0.9kb. Lane 6&9: VF2 and VR were used to amplify water. As expected, nothing was amplified. Lane 7:The rep region was amplified correctly. I expected a band at 3.352kb and the band occurred in this region.
Discussion
- I did some trouble shooting on this PCR with advice from: http://www.med.yale.edu/genetics/ward/tavi/Trblesht.html.
- In most cases, I just got as close to the recommended annealing temperature and extension time in order to decrease the amount of unspecific products.
- I only ran 30 cycles as opposed to 35 in the past. I think this has made the efficiency of my taq more consistent, especially with the longer products.
- I also added an additional 10 seconds to last 20 cycles of the rep region, http://www.med.yale.edu/genetics/ward/tavi/Trblesht.html, recommends this extra time because the taq supposedly loses efficiency and thus there will be multiple products of suspicious size. This seems to have worked for the rep region, which contains only one product.
Transformation
- Margaret
- pSMC121 (we were running out of template)
- re-streaked transformants on 8-5. There was a lawn of transformants.
Re-streak Library
- Margaret
- pSB1A7, L51020, psb3k3, psb1a3, oriT, psb1a2, db3.1 strain, i14032
Construction of p+r, g+t
- Grace
- Gel purified nir1, GFPf1, GFP, E0240, I14032, slr1, slr2, pilA PCR products
- RE digest
- nir, GFP, GFPf, I14032 digested with EcoRI and SpeI
- E0240 digested with EcoRI and PstI
- slr1, slr2, pilA digested with PstI and XbaI
- Ran RE digests on gel
- Incomplete digestion of nir, slr1, slr2, pilA
- Krystle
- Gel purified RE digests of nir1, GFPf1, GFP, E0240, I14032, slr1, slr2, pilA
- Ligated at 37C overnight:
- nir + B0030
- I14032 + B0030
- GFP + B0015
- GFPf + B0015
- Supposed to ligate at 25C
PCR
- Grace
- Redid Saturday's PCR of BB-pRl1383a and pRL1383a alongside test of PCR contamination
- Ran on 2.5% gel for 1 hour at 95V
- Gel doesn't match that from Saturday
- Redid PCR again to verify Saturday/today's PCR
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"