Team:BCCS-Bristol/Calendar-Notebook/6 August 2008
From 2008.igem.org
Tgorochowski (Talk | contribs) |
|||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html><link rel="stylesheet" href="http://homepage.mac.com/tgorochowski/iGEM/bccs-igem.css" type="text/css"></html> | ||
| + | __NOTOC__ | ||
| + | <div class="bccsNavBar"> | ||
| + | {| align="center" | ||
| + | !align="center"|[[Team:BCCS-Bristol|Home]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Team|The Team]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Project|The Project]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Parts|Submitted Parts]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Modeling|Modelling]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Notebook|Wet Lab]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Calendar|Calendar]] | ||
| + | !align="center"|[[Team:BCCS-Bristol/Misc|Miscellaneous]] | ||
| + | |} | ||
| + | <br> | ||
| + | </div> | ||
| + | |||
==BioBrick Transformation== | ==BioBrick Transformation== | ||
| Line 13: | Line 29: | ||
|} | |} | ||
| - | {| align="center" | + | {| align="center" border=1 |
| | | | ||
L= HyperLadder I (BIOLINE) | L= HyperLadder I (BIOLINE) | ||
N= Negative control | N= Negative control | ||
| + | |||
| + | Numbers= See text | ||
|} | |} | ||
The transformation efficiency of the electroportation was significant higher than of the chemical competent cells. Therefore, an own stock of electric competent cells was made. | The transformation efficiency of the electroportation was significant higher than of the chemical competent cells. Therefore, an own stock of electric competent cells was made. | ||
| - | |||
==Agar plug assay== | ==Agar plug assay== | ||
TR235 (E. coli with construct to detect adhesion to hydrophobic surfaces) arrived from Princeton (Thomas J. Silhavy). Cells are viable, growth curve done. 11µm PS beads shown to be moved by bacteria (MG1655); we observed ‘jiggling’ of beads when bacteria present (not seen in no-bacteria control). Also, we observed significant movement in different directions (distances of approx 1x to 5x length of the beads by eye) of beads that were close together, suggesting that movement was not due to currents in the medium. We saw no distance covered in almost all the control beads, a few moved small distances, so some currents are present. Bacteria and beads were left together for ~30mins to allow time for adhesion. No movement has yet been seen when the bacteria and beads are not left to adhere. We will look again at this, as we may have not previously looked closely enough (photographing over a time course of a few minutes). | TR235 (E. coli with construct to detect adhesion to hydrophobic surfaces) arrived from Princeton (Thomas J. Silhavy). Cells are viable, growth curve done. 11µm PS beads shown to be moved by bacteria (MG1655); we observed ‘jiggling’ of beads when bacteria present (not seen in no-bacteria control). Also, we observed significant movement in different directions (distances of approx 1x to 5x length of the beads by eye) of beads that were close together, suggesting that movement was not due to currents in the medium. We saw no distance covered in almost all the control beads, a few moved small distances, so some currents are present. Bacteria and beads were left together for ~30mins to allow time for adhesion. No movement has yet been seen when the bacteria and beads are not left to adhere. We will look again at this, as we may have not previously looked closely enough (photographing over a time course of a few minutes). | ||
| + | |||
| + | |||
| + | {| align="center" | ||
| + | | | ||
| + | [[Image:BeadMovment1.JPG | 300px]] | ||
| + | [[Image:BeadMovment2.JPG | 300px]] | ||
| + | [[Image:BeadMovment3.JPG | 300px]] | ||
| + | |} | ||
| + | {| align="center" | ||
| + | | | ||
| + | !align="center" width="10%"| | ||
| + | !align="center" width="10%"|0 mins | ||
| + | !align="center" width="25%"| | ||
| + | !align="center" width="10%"|1 min | ||
| + | !align="center" width="25%"| | ||
| + | !align="center" width="10%"|2 mins | ||
| + | !align="center" width="10%"| | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| align="center" | ||
| + | | | ||
| + | !align="center" width="10%"| | ||
| + | !align="center" width="80%"|'''Circles represent position at 0 mins, one colour for each bead. For example, the bead that starts in red circle moves down and right, while that in the yellow circle moves left.''' | ||
| + | !align="center" width="10%"| | ||
| + | |} | ||
| + | |||
Latest revision as of 09:58, 14 September 2008
BioBrick Transformation
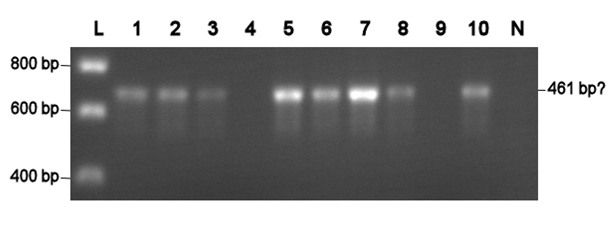
Last Thursday, the primers [http://partsregistry.org/wiki/index.php/Part:BBa_G00100 VF2] and [http://partsregistry.org/Part:BBa_G00101 VR] arrived. These primers allow a confirmation of a succesful transformation of most BioBrick parts by means of PCR. The resulting fragment length depends on the specific BioBrick DNA.
Using VF2 and VR, a colony PCR are was conducted with all colonies that were obtained with chemical competent cells so far. Two colonies with the [http://partsregistry.org/Part:BBa_J63005 yeast ADH1 promoter] (No. 1 and 2, see left photo) and three colonies with a [http://partsregistry.org/Part:BBa_E0240 GFP generator] (No. 3-5, see left photo) were identified as positive. Thus, all colonies are positive, but the transformation efficiency is too low, since the result was only one or zero colonies per transformation attempt.
Electroporation should give higher efficiencies. An attempt with a new BioBrick ([http://partsregistry.org/Part:BBa_J63002 ADH1 terminator]) resulted in 69 colonies! Ten of them were analysed using PCR. The resulting fragment should have a length of 461 bp, but from the eigth positive colonies fragments between 650-750 bp were obtained (see right photo). This is probably due to a mutation which might result in a different binding location for one of the primers. Another explanation might be that the denoted length is wrong. Since this BioBrick is not important for our project, it will be disregarded.
|
L= HyperLadder I (BIOLINE) N= Negative control Numbers= See text |
The transformation efficiency of the electroportation was significant higher than of the chemical competent cells. Therefore, an own stock of electric competent cells was made.
Agar plug assay
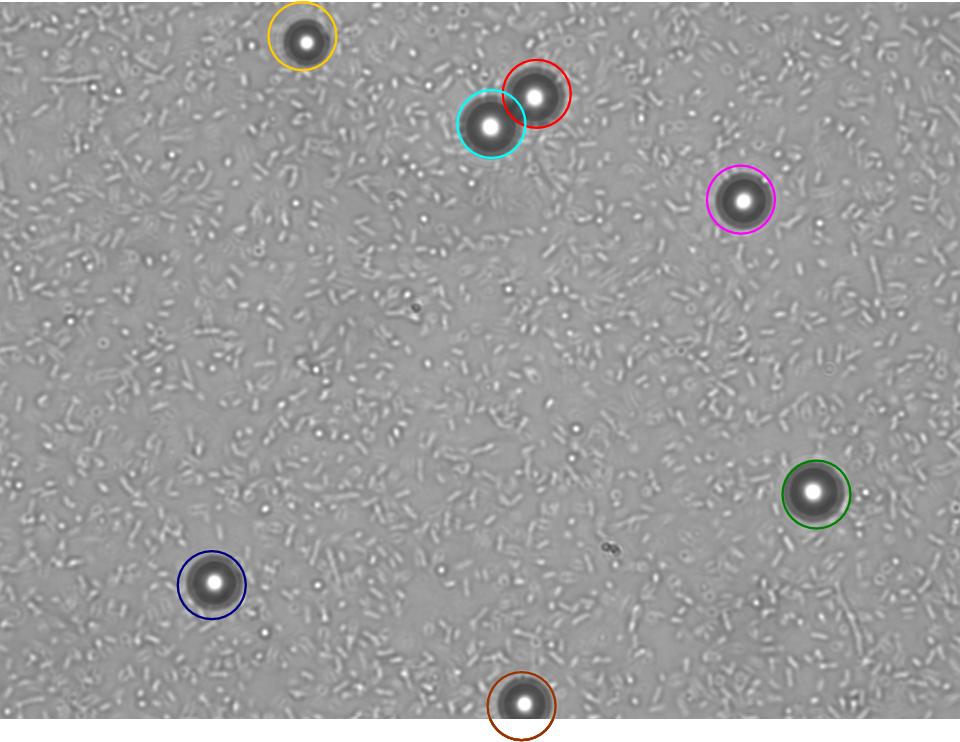
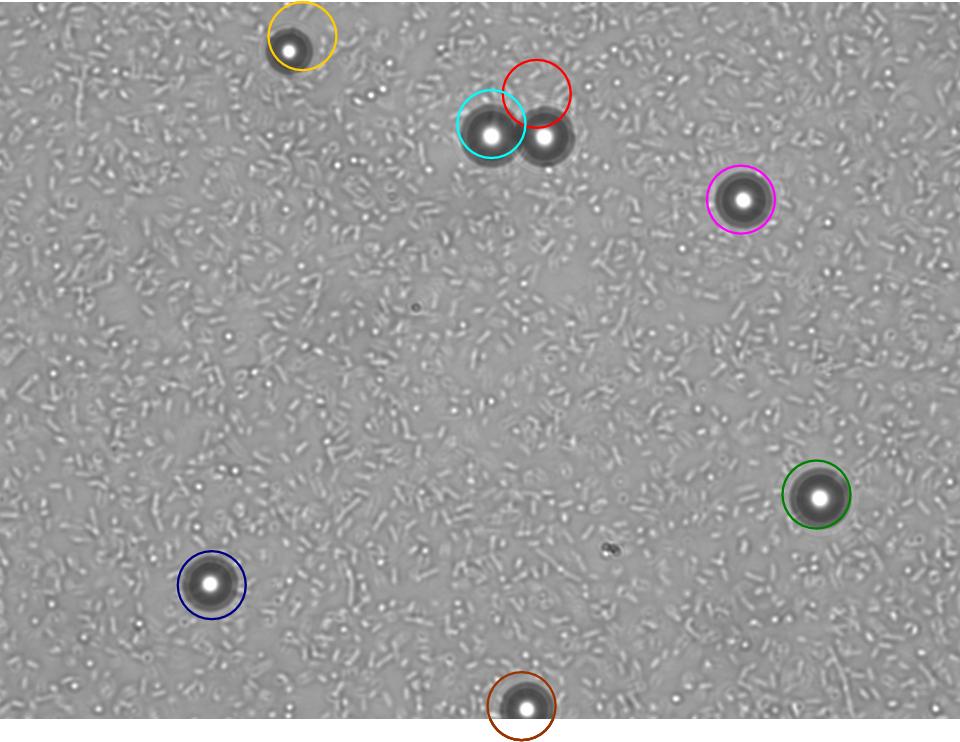
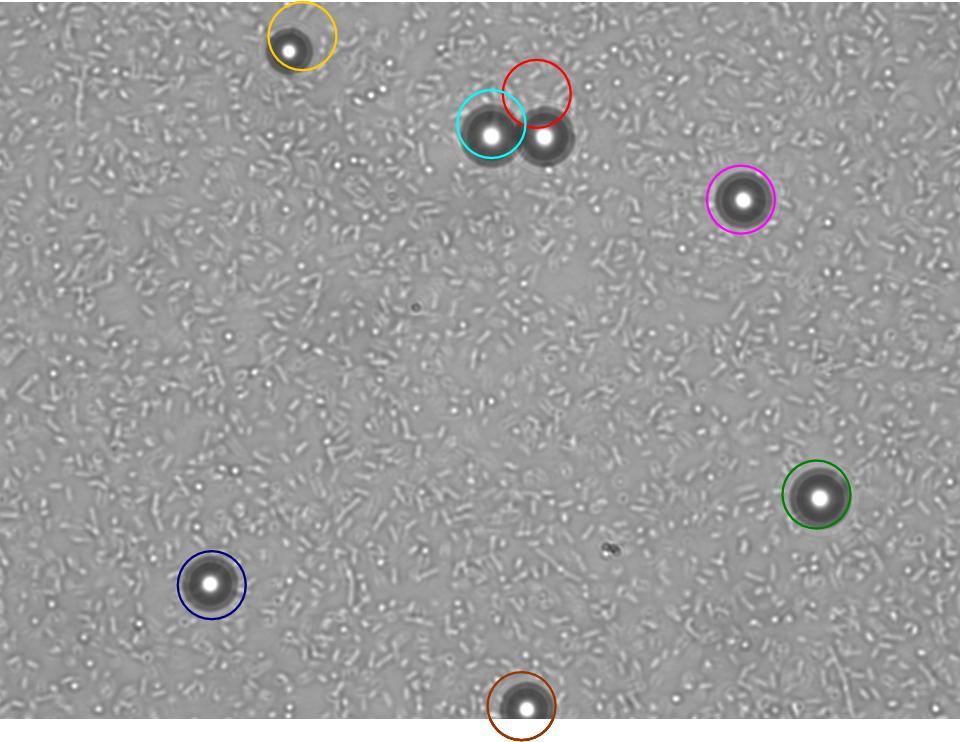
TR235 (E. coli with construct to detect adhesion to hydrophobic surfaces) arrived from Princeton (Thomas J. Silhavy). Cells are viable, growth curve done. 11µm PS beads shown to be moved by bacteria (MG1655); we observed ‘jiggling’ of beads when bacteria present (not seen in no-bacteria control). Also, we observed significant movement in different directions (distances of approx 1x to 5x length of the beads by eye) of beads that were close together, suggesting that movement was not due to currents in the medium. We saw no distance covered in almost all the control beads, a few moved small distances, so some currents are present. Bacteria and beads were left together for ~30mins to allow time for adhesion. No movement has yet been seen when the bacteria and beads are not left to adhere. We will look again at this, as we may have not previously looked closely enough (photographing over a time course of a few minutes).
| 0 mins | 1 min | 2 mins |
|---|
| Circles represent position at 0 mins, one colour for each bead. For example, the bead that starts in red circle moves down and right, while that in the yellow circle moves left. |
|---|
Chemotaxis assay (Yu, H.S. et al. (1997) FEMS Microbiology Letters, 156, 265-269.) has not worked so far. Currents can be seen flowing from the aspartate plug, pushing bacteria away. The focus of this week will be on getting this fixed, with a view to combining chemotaxis and the ability to move beads, allowing us to move beads in a particular direction.
The GRN for switching on the chemotactic response by binding to a bead, and signaling to nearby bacteria that a particle is in the vicinity, is almost finalised.
 "
"