Team:Caltech/Project/Oxidative Burst
From 2008.igem.org
| (16 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
__NOTOC__ | __NOTOC__ | ||
==Introduction== | ==Introduction== | ||
| - | In order to help guard against infections of the gut, we wish to engineer a strain of E. coli capable of killing bacterial pathogens. White blood cells called neutrophils are already very efficient at killing bacteria. Neutrophils kill bacteria by | + | In order to help guard against infections of the gut, we wish to engineer a strain of ''E. coli'' capable of killing bacterial pathogens. White blood cells called neutrophils are already very efficient at killing bacteria. Neutrophils kill bacteria by bombarding them with a variety of reactive oxygen species including superoxide, hydrogen peroxide and hydrochlorous acid. The reactive oxygen species kill the bacteria by shredding biological molecules with their potent oxidizing properties. However, neutrophils do not migrate to the large intestinal lumen where pathogens can reside. Because bacteria are well adapted to live in the gut, this project’s goal is to engineer a strain of ''E. coli'' to detect and kill invading bacterial pathogens by means of a sudden burst of hydrogen peroxide. |
===Detection=== | ===Detection=== | ||
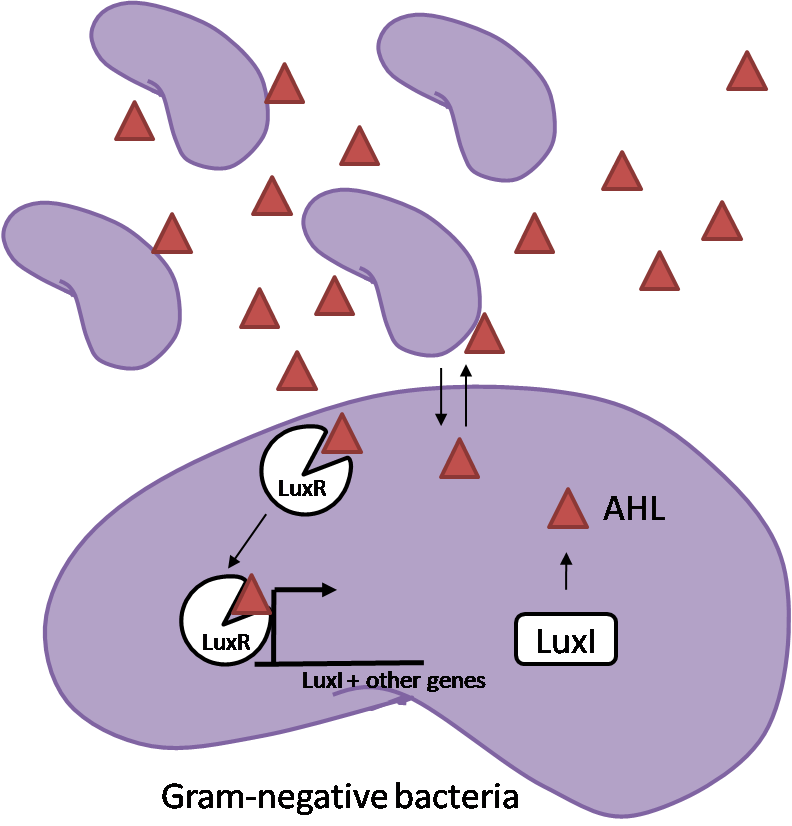
[[Image:Quorum_sensing.png|thumb|left|Figure 1 - Quorum sensing in gram negative bacteria]] | [[Image:Quorum_sensing.png|thumb|left|Figure 1 - Quorum sensing in gram negative bacteria]] | ||
| - | Bacteria are able to communicate between individuals of the same species by way of quorum sensing. Small molecules serve as the signal between individual cells. Gram negative bacteria use acylhomoserine lactones (AHL), which can freely diffuse across the cell membrane. The quorum sensing machinery relies in two enzymes, LuxI, an AHL producer, and LuxR, an AHL-dependent transcriptional activator. Figure 1 illustrates how the quorum sensing system works. In isolation, each bacterium constitutively produces a small amount of AHL, which quickly diffuses into the surroundings. If other bacteria of the same species are also nearby, the AHL will diffuse across their membrane where it will bind to LuxR. LuxR activates transcription of several genes, including luxI. A positive feedback loop is created, in which more AHL induces more LuxI, which in turn produces more AHL. Each species of gram negative bacteria produces a unique AHL, requiring unique LuxI and LuxR proteins, and so avoids crosstalk between species. A group of bacteria can thus toggle between an “off” state and an “on” state by using quorum sensing. | + | Bacteria are able to communicate between individuals of the same species by way of quorum sensing. Small molecules serve as the signal between individual cells. Gram negative bacteria use acylhomoserine lactones (AHL), which can freely diffuse across the cell membrane. The quorum sensing machinery relies in two enzymes, LuxI, an AHL producer, and LuxR, an AHL-dependent transcriptional activator. Figure 1 illustrates how the quorum sensing system works. In isolation, each bacterium constitutively produces a small amount of AHL, which quickly diffuses into the surroundings. If other bacteria of the same species are also nearby, the AHL will diffuse across their membrane where it will bind to LuxR. LuxR activates transcription of several genes, including ''luxI''. A positive feedback loop is created, in which more AHL induces more LuxI, which in turn produces more AHL. Each species of gram negative bacteria produces a unique AHL, requiring unique LuxI and LuxR proteins, and so avoids crosstalk between species. A group of bacteria can thus toggle between an “off” state and an “on” state by using quorum sensing. |
| - | + | Our engineered strain will not participate directly in quorum sensing, but instead will eavesdrop on the conversation. It will be engineered to constitutively express a LuxR able to detect a species' AHL. By taking advantage of the specificity of quorum sensing, our engineered strain will be able to be tuned to specifically respond to a variety of bacterial pathogens. Quorum sensing has been shown to be associated with the expression of virulence genes in several enteric pathogens, including ''Vibrio cholerae'' <sup>1</sup>. Once the AHL is bound, LuxR will activate a set of genes which will lead to the overproduction of hydrogen peroxide, killing the invading cell. | |
| - | + | ||
| - | Our engineered strain will not participate directly in quorum sensing, but instead will eavesdrop on the conversation. It will be engineered to constitutively express a LuxR able to detect a species AHL. By taking advantage of the specificity of quorum sensing, our engineered strain will be able to be tuned to specifically respond to a variety of bacterial pathogens. Once the AHL is bound, LuxR will activate a set of genes which will lead to the overproduction of hydrogen peroxide, killing the invading cell. | + | |
===Response=== | ===Response=== | ||
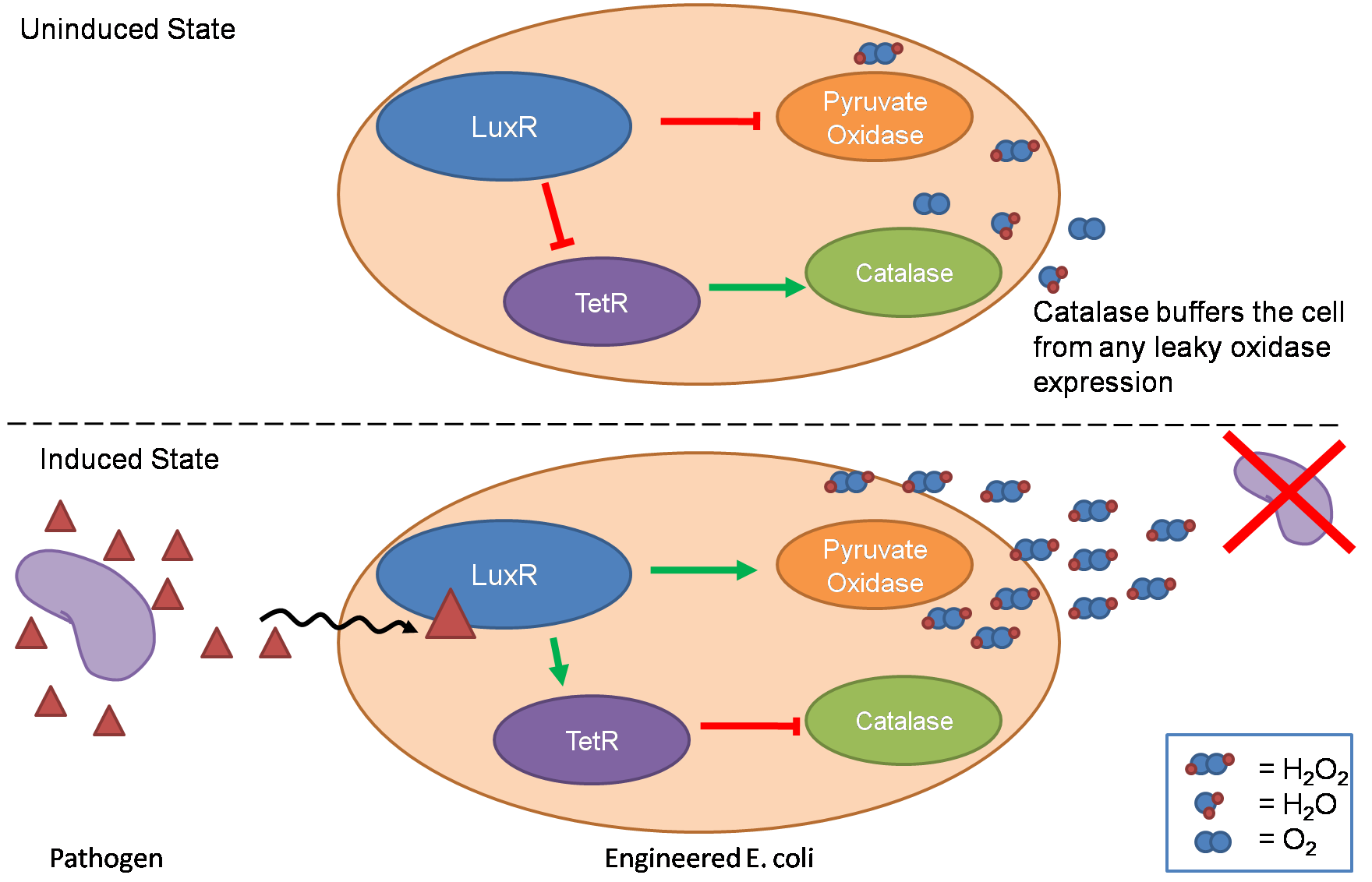
| - | + | [[Image:Oxidative burst states.png|400px|thumb|right|Figure 2 - Detection scheme used by our engineered ''E. coli'']] | |
| - | + | After sensing the presence of an invading pathogen, we want to engineer our ''E. coli'' to produce lethal amounts of hydrogen peroxide relatively quickly. We are not concerned with having the engineered ''E. coli'' survive, as it is reasonable to assume that there are “unactivated” cells far away from the pathogen that could sustain the population. Even if all of the oxidative burst cells were wiped out, the population could be regenerated. In combination with the [[Team:Caltech/Project/Population_Variation|<font style="color:#BB4400">population variation</font>]] subproject, the "master cell line" would differentiate into the oxidative burst state, thus reseeding the population. After LuxR binds AHL, it will activate transcription of pyruvate oxidase, which uses pyruvate to produce hydrogen peroxide in the following reaction <sup>2</sup>: | |
| - | + | Pyruvate + phosphate + O<sub>2</sub> --> H<sub>2</sub>O<sub>2</sub> + CO<sub>2</sub> + acetyl phosphate | |
| - | Even though our activated engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough to be cytotoxic and then fissle, killing only themselves and little else. To avoid this problem, an ''E. coli'' catalase will be | + | ''Streptococcus pneumoniae'' naturally uses pyruvate oxidase to kill off competing bacteria when infecting the lungs. Having already demonstrated its activity and antibacterial properties ''in vivo'', pyruvate oxidase was an attractive oxidase with which to work. |
| + | |||
| + | Even though our activated engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough H<sub>2</sub>O<sub>2</sub> to be cytotoxic and then fissle, killing only themselves and little else. To avoid this problem, an ''E. coli'' catalase will be expressed by default, and then turned off shortly after pyruvate oxidase expression is triggered. We are accomplishing this regulation by putting ''katG'' (on of two ''E. coli'' catalase genes) behind the TetR sensitive promoter (pTet) and having TetR co-transciptionally expressed with the oxidase. The time it takes for TetR to accumulate in the cell provides the delay in repressing KatG expression. To ensure KatG is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted. The final strain will have deletions of both catalases, ensuring that we have complete control over catalase expression. | ||
===System Design=== | ===System Design=== | ||
| + | <gallery widths="375px" heights="250px" > | ||
| + | Image:Oxidative_burst_system_design.png|Figure 3 - System design for the oxidative burst. | ||
| + | Image:Oxidative_burst_system_response.png|Figure 4 - Ideal system response for the engineered E. coli after being exposed to AHL. | ||
| + | </gallery> | ||
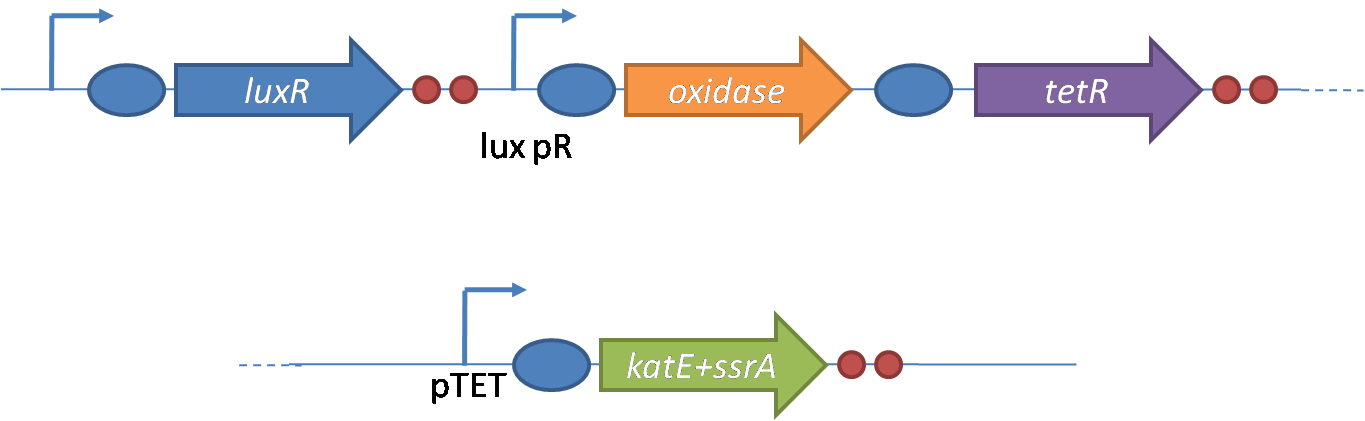
| - | + | The entire system will be engineered onto a high copy plasmid in ''E. coli'' as seen in Figure 3. Promoters are shown as bent, thin arrows, genes as thick arrows, ribosome binding sites as blue ovals, and a double transcriptional terminator as two red circles. Initially, the system will function independently, with ''luxR'' under a constitutive promoter. Later, ''luxR'' will be put under the control of a recombinase system that will only turn on the pathway randomly in a subset of cells. This will be one of three fates the master strain will be able to differentiate into. For more information, visit the [[Team:Caltech/Project/Population_Variation|<font style="color:#BB4400">population variation</font>]] page. | |
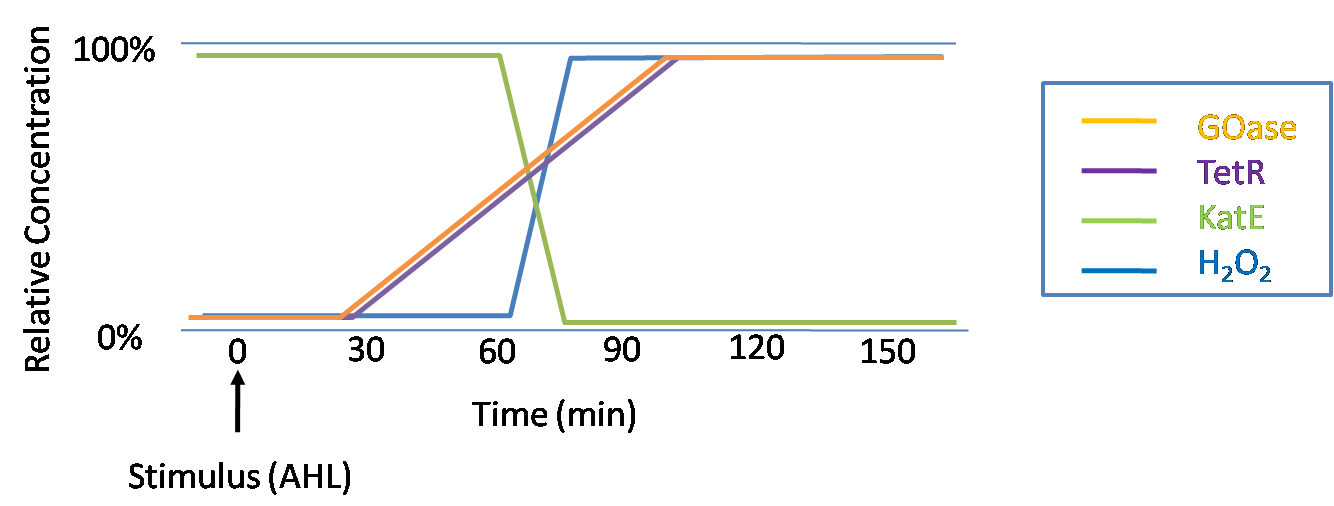
| - | + | An ideal system would behave as outlined in Figure 4. The figure shows the relative abundances of the proteins and molecules used in the system. It is only meant to show if a particular species is relatively high or low, not its absolute concentration. | |
| - | |||
| - | |||
| - | |||
==Results== | ==Results== | ||
===Constructs=== | ===Constructs=== | ||
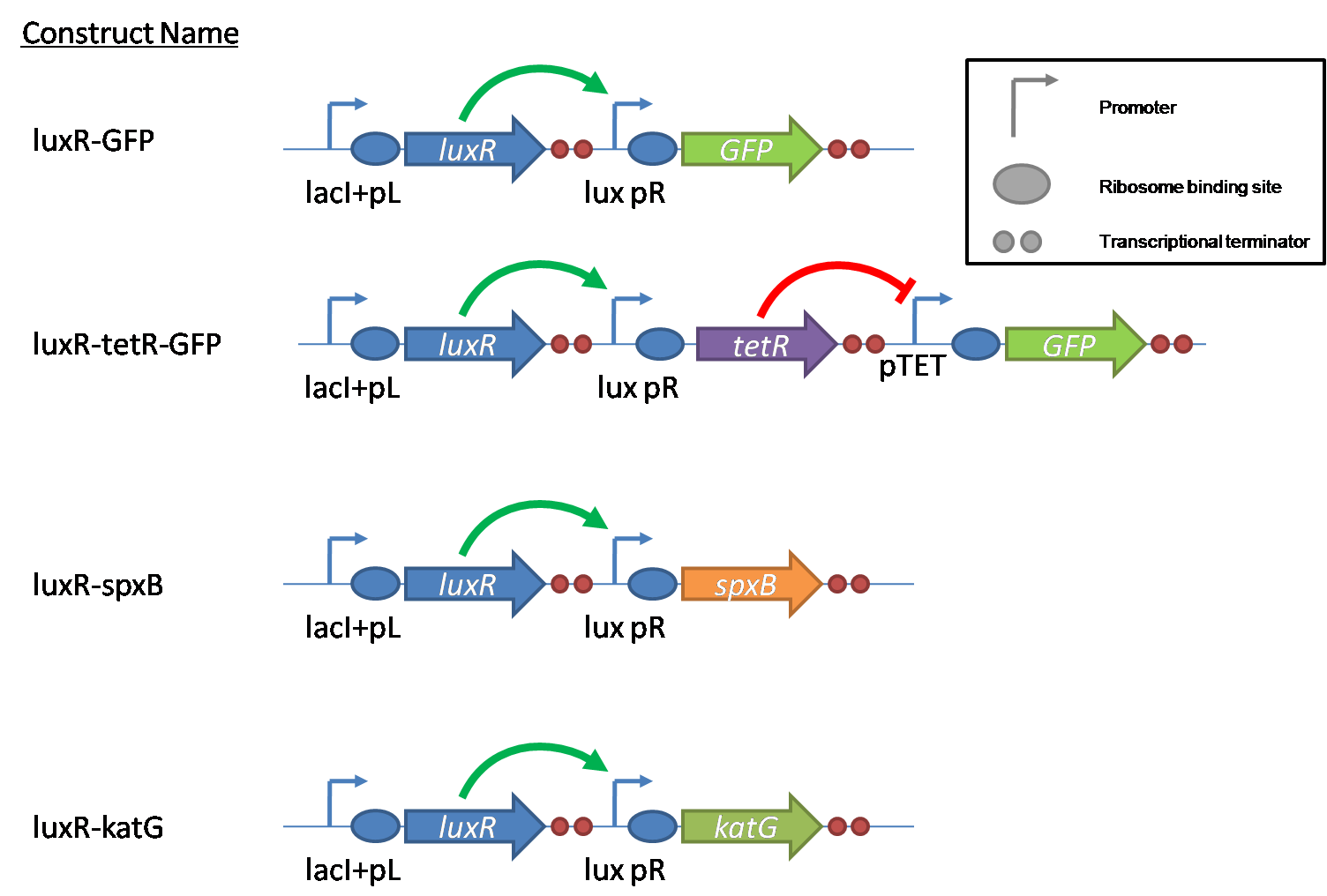
| - | [[Image:Igem constructs.png|thumb| | + | [[Image:Igem constructs.png|thumb|right|400px|Figure 5 - Constructs made to characterize the performance of SpxB and KatG, as well as their genetic control elements.]] |
| - | + | The constructs shown in figure 5 were made using the standard assembly method. They were made to test the various regulatory elements and the functionality of the oxidase and catalase. | |
| - | The constructs shown in figure | + | {{Clear}} |
===Characterization of LuxR Inducer=== | ===Characterization of LuxR Inducer=== | ||
| - | [[Image:LuxR TetR response curves.png|thumb|left|405px|box|Figure | + | [[Image:LuxR TetR response curves.png|thumb|left|405px|box|Figure 6 - Characterization of LuxR inducer and TetR inverter by flow cytometry. The figure shows the fluorescence of GFP in the constructs luxR-GFP and luxR-tetR-GFP. The left axis corresponds to the LuxR data and the right axis corresponds to the TetR data. At 100 nM AHL, cells with the luxR-GFP construct grew very poorly.]] |
| - | The luxR inducer was characterized using the luxR-GFP construct. GFP provides a relative gauge of expression levels of SpxB in the final construct. The cultures were grown over a range of AHL concentrations and analyzed by flow cytometery. Figure | + | The luxR inducer was characterized using the luxR-GFP construct. GFP provides a relative gauge of expression levels of SpxB in the final construct. The cultures were grown over a range of AHL concentrations and analyzed by [[Team:Caltech/Protocols/Flow_cytometry|<font style="color:#BB4400">flow cytometery</font>]]. Figure 6 shows that the LuxR inducer behaves as expected. The system shows tight repression in the uninduced state, and begins to show significant expression at 1nM AHL. There is a 50 fold difference between the uninduced and fully induced (10nM AHL) state. The sudden drop at 100nM AHL is explained by the fact that these cells grew very poorly, possibly the result of toxic proteins loads. The LuxR inducer functions well if the concentration of AHL is kept below 10nM. |
| + | {{Clear}} | ||
===Characterization of TetR Inverter=== | ===Characterization of TetR Inverter=== | ||
| - | The TetR inverter was characterized by using a luxR-tetR-GFP construct. In this context, GFP provides a relative gauge of expression levels of KatG in the final construct. Figure | + | The TetR inverter was characterized by using a luxR-tetR-GFP construct. In this context, GFP provides a relative gauge of expression levels of KatG in the final construct. Figure 6 shows that the tetracycline inverter behaved as expected. At high concentrations of AHL, the TetR inverter shows near full repression, as compared to the lower concentrations of AHL. Similar to the LuxR inducer, the TetR inverter showed a 50 fold difference in expression between the uninduced (low AHL) and induced (high AHL) states. However the absolute maximum level of expression from the TetR inverter was much lower than that from the LuxR inducer (compare the scale of the left TetR axis to the scale of the left LuxR axis). The low expression levels can be explained by the leakiness of the LuxR promoter, resulting in a basal level of repression from the TetR inverter. |
| - | The low expression level is advantageous for the engineered E. coli because catalase is a very efficient scavenger of hydrogen peroxide. The low expression level ensures there is enough KatG to buffer any leaky pyruvate oxidase expression, yet still clear quickly from the cytoplasm. Additionally, in the uninduced state there should be extremely low levels of KatG. The LAA degradation tag should reduce KatG levels even further. | + | The low expression level is advantageous for the engineered ''E. coli'' because catalase is a very efficient scavenger of hydrogen peroxide. The low expression level ensures there is enough KatG to buffer any leaky pyruvate oxidase expression, yet still clear quickly from the cytoplasm. Additionally, in the uninduced state there should be extremely low levels of KatG. The LAA degradation tag should reduce KatG levels even further. |
| - | === | + | ===Pyruvate Oxidase Produces Hydrogen Peroxide ''in vivo''=== |
| - | [[Image:H2O2 timecourse.png|thumb|right|400px|Timecourse of | + | [[Image:H2O2 timecourse.png|thumb|right|400px|Figure 7: Timecourse of H<sub>2</sub>O<sub>2</sub> production in JI377 cells with the luxR-spxB construct in SOC. JI377 cells were back diluted 1:100 and grown to an OD600 of ~0.8. Hydrogen peroxide in the supernatant was assayed every half hour. The arrow indicates induction with AHL at time 0 hr. Error bars represent one standard deviation(n=3) and are too small to be visible on the negative control.]] |
| - | JI377 cells expressing pyruvate oxidase behind the LuxR inducer were able to produce significant amounts of hydrogen peroxide when cultured in rich media, reaching a maximum of 800 µM | + | Using the [[Team:Caltech/Protocols/H2O2_Production_Assay|<font style="color:#BB4400">H<sub>2</sub>O<sub>2</sub> production assay protocol</font>]], JI377 (Δ''ahpCF'kan::'ahpF''Δ(''katG17''::Tn''10'') ''katE12''::Tn''10'') cells expressing pyruvate oxidase behind the LuxR inducer were able to produce significant amounts of hydrogen peroxide when cultured in rich media, reaching a maximum of 800 µM H<sub>2</sub>O<sub>2</sub> in 2 hours (see figure 7).This is significant because the engineered cells can produce more than the minimum inhibitory concentration (MIC) for hydrogen peroxide of some human pathogens <sup>2</sup>. The decline in hydrogen peroxide concentration after the peak is explained by the reaction of hydrogen peroxide with the SOC media, since H<sub>2</sub>O<sub>2</sub> has a half life of about 3.5 hours in SOC (data not shown). The most surprising finding was that the cells began expressing hydrogen peroxide a full hour before they were induced with AHL. The current hypothesis is that some component of the rich SOC media mimicked AHL and activated LuxR. Uninduced expression from LuxR was also observed when culturing cell transformed with the luxR-GFP construct in LB media, as cell pellets were visibly green. However, no such autoinduction occurred in the defined minimal media M9. It would be interesting to see if LuxR spontaneously activates expression in a rich defined media, such as Neidhardt. However, the significant finding is that pyruvate oxidase is capable of producing bacteriostatic amounts of hydrogen peroxide in a short amount of time in vivo. |
===Functionality of KatG=== | ===Functionality of KatG=== | ||
| - | [[Image:KatG MIC.png|thumb|left| | + | [[Image:KatG MIC.png|thumb|left|380px| KatG +LAA confers hydrogen peroxide resistance. Saturated cultures of SN0037 cells with the construct luxR-katG were backdiluted 1:50 into SOC with or without AHL (0 or 100nM respectively) and various concentrations of hydrogen peroxide. The minimum inhibitory concentrations (MIC) for each condition are plotted.]] |
| - | Functional expression of KatG from the luxR-katG construct protects cells from hydrogen peroxide. Figure 5 shows that expression of the KatG +LAA degradation tag in catalase deficient cells ( | + | Functional expression of KatG from the luxR-katG construct protects cells from hydrogen peroxide. Figure 5 shows that expression of the KatG +LAA degradation tag in catalase deficient cells (SN0037, Δ''katG''Δ''katE'') increases their [[Team:Caltech/Protocols/MIC_Assay|<font style="color:#BB4400">measured</font>]] minimum inhibitory concentration (MIC) by an order of magnitude. It is significant in that the MIC range of 3.0 - 10.0 mM H<sub>2</sub>O<sub>2</sub> is well above to the MIC value of wild type E. coli (MIC = 0.8 uM). Even with a degradation tag, KatG is still functional and is sufficient to protect the cell from exogenous hydrogen peroxide. In an uninduced state, KatG will be able to buffer the engineered cell from leaky hydrogen peroxide production and should be degraded on the order of minutes because of the LAA tag. However in the final engineered strain, KatG will be expressed from behind the tetracycline inverter, not the LuxR system. Further tests will be necessary to ensure that the reduced expression levels from tetracycline inverter still produce enough KatG to raise the MIC of SN0037. The important result is that KatG +LAA is functionally expressed and is able to raise the MIC of SN0037. |
| + | {{Clear}} | ||
====Coculture Assay==== | ====Coculture Assay==== | ||
| - | |||
[[Image:Coculture table.png|thumb|right|400px|Table 1 - CFU counts of a competing E. coli strain before and after co-culturing with the engineered strain. Target JI377 strain (AmpR KanR) was inoculated in various amounts (A, B and C) into 2.5 mL SOC of engineered JI377 strain (AmpR). Production of hydrogen peroxide was induced with 10 nM AHL upon target strain inoculation. Cultures were serially diluted and plated onto LB+Kan at the given times. Key: +++ = lawn; ++ = many colonies, + = few colonies, - = no colonies.]] | [[Image:Coculture table.png|thumb|right|400px|Table 1 - CFU counts of a competing E. coli strain before and after co-culturing with the engineered strain. Target JI377 strain (AmpR KanR) was inoculated in various amounts (A, B and C) into 2.5 mL SOC of engineered JI377 strain (AmpR). Production of hydrogen peroxide was induced with 10 nM AHL upon target strain inoculation. Cultures were serially diluted and plated onto LB+Kan at the given times. Key: +++ = lawn; ++ = many colonies, + = few colonies, - = no colonies.]] | ||
| - | Finally, we wanted to see if the engineered JI377 cells could directly kill or inhibit the growth of a competing JI377 strain (due to time constraints, only JI377 with a luxR-spxB construct could be tested). We carried out co-culture assays in which varying amounts of the target strain were introduced to a heavy (OD600 of ~0.8) culture of the engineered strain, and incubated the co-culture for 6 hours after induction with AHL. Introducing the target strain to a dense population of our engineered E. coli was meant to mimic the situation in the gut where in invading pathogen will face an already established culture of engineered E. coli. Survival of the target strain was assayed by a colony forming unit (CFU) count both immediately upon introduction and after 6 hours of co-culturing. Table 1 summarizes the results. The CFU data show that the engineered strain was able to kill the target JI377 strain, while the target strain was unaffected or was able to grow in co-cultures with the negative control (JI377 with no pyruvate oxidase). | + | Finally, we wanted to see if the engineered JI377 cells could directly kill or inhibit the growth of a competing JI377 strain (due to time constraints, only JI377 with a luxR-spxB construct could be tested). We carried out [[Team:Caltech/Protocols/Coculture_Inhibition_Assay|<font style="color:#BB4400">co-culture assays</font>]] in which varying amounts of the target strain were introduced to a heavy (OD600 of ~0.8) culture of the engineered strain, and incubated the co-culture for 6 hours after induction with AHL. Introducing the target strain to a dense population of our engineered ''E. coli'' was meant to mimic the situation in the gut where in invading pathogen will face an already established culture of engineered ''E. coli''. Survival of the target strain was assayed by a colony forming unit (CFU) count both immediately upon introduction and after 6 hours of co-culturing. Table 1 summarizes the results. The CFU data show that the engineered strain was able to kill the target JI377 strain, while the target strain was unaffected or was able to grow in co-cultures with the negative control (JI377 with no pyruvate oxidase). |
| - | + | ||
==Conclusion== | ==Conclusion== | ||
| - | We have successfully demonstrated the overproduction of hydrogen peroxide in E. coli (JI377) using the pyruvate oxidase from | + | We have successfully demonstrated the overproduction of hydrogen peroxide in ''E. coli'' (JI377) using the pyruvate oxidase from ''S. pneumoniae''. This “oxidative burst” is sufficient to kill a competing strain of JI377 cells in co-culture assays. A genetic circuit was also constructed to regulate expression of pyruvate oxidase and catalase in response to the quorum sensing molecule acylhomoserine lactone. AHL and other quorum sensing molecules have been shown to be associated with the production of virulence factors of enteric pathogens, most notably ''Vibrio cholerae'' <sup>1</sup>. Flow cytometry experiments showed that in a defined minimal media, the control circuit behaves as expected over a wide range of AHL concentrations. However in richer medias, the LuxR inducer is able to spontaneously turn on. The cause of this is still unknown, but it is possible some molecule in the undefined media closely mimics AHL. Overall, the data shows that engineered ''E. coli'' are capable of producing sufficient hydrogen peroxide to kill competing bacteria. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===References=== | ===References=== | ||
| - | + | # Kaper, J.B., and Sperandio, V. (2005) ''Infection and Immunity'' '''73''', 3197-3209 | |
| + | # Pericone, C.D, Park, S. Imlay, J.A, and Weiser, J.N. (2003) ''Journal of Bacteriology'' '''185''', 6815-6825 | ||
}} | }} | ||
Latest revision as of 20:23, 25 October 2008
|
People
|
Pathogen Defense by Oxidative Burst
IntroductionIn order to help guard against infections of the gut, we wish to engineer a strain of E. coli capable of killing bacterial pathogens. White blood cells called neutrophils are already very efficient at killing bacteria. Neutrophils kill bacteria by bombarding them with a variety of reactive oxygen species including superoxide, hydrogen peroxide and hydrochlorous acid. The reactive oxygen species kill the bacteria by shredding biological molecules with their potent oxidizing properties. However, neutrophils do not migrate to the large intestinal lumen where pathogens can reside. Because bacteria are well adapted to live in the gut, this project’s goal is to engineer a strain of E. coli to detect and kill invading bacterial pathogens by means of a sudden burst of hydrogen peroxide. DetectionBacteria are able to communicate between individuals of the same species by way of quorum sensing. Small molecules serve as the signal between individual cells. Gram negative bacteria use acylhomoserine lactones (AHL), which can freely diffuse across the cell membrane. The quorum sensing machinery relies in two enzymes, LuxI, an AHL producer, and LuxR, an AHL-dependent transcriptional activator. Figure 1 illustrates how the quorum sensing system works. In isolation, each bacterium constitutively produces a small amount of AHL, which quickly diffuses into the surroundings. If other bacteria of the same species are also nearby, the AHL will diffuse across their membrane where it will bind to LuxR. LuxR activates transcription of several genes, including luxI. A positive feedback loop is created, in which more AHL induces more LuxI, which in turn produces more AHL. Each species of gram negative bacteria produces a unique AHL, requiring unique LuxI and LuxR proteins, and so avoids crosstalk between species. A group of bacteria can thus toggle between an “off” state and an “on” state by using quorum sensing. Our engineered strain will not participate directly in quorum sensing, but instead will eavesdrop on the conversation. It will be engineered to constitutively express a LuxR able to detect a species' AHL. By taking advantage of the specificity of quorum sensing, our engineered strain will be able to be tuned to specifically respond to a variety of bacterial pathogens. Quorum sensing has been shown to be associated with the expression of virulence genes in several enteric pathogens, including Vibrio cholerae 1. Once the AHL is bound, LuxR will activate a set of genes which will lead to the overproduction of hydrogen peroxide, killing the invading cell. ResponseAfter sensing the presence of an invading pathogen, we want to engineer our E. coli to produce lethal amounts of hydrogen peroxide relatively quickly. We are not concerned with having the engineered E. coli survive, as it is reasonable to assume that there are “unactivated” cells far away from the pathogen that could sustain the population. Even if all of the oxidative burst cells were wiped out, the population could be regenerated. In combination with the population variation subproject, the "master cell line" would differentiate into the oxidative burst state, thus reseeding the population. After LuxR binds AHL, it will activate transcription of pyruvate oxidase, which uses pyruvate to produce hydrogen peroxide in the following reaction 2: Pyruvate + phosphate + O2 --> H2O2 + CO2 + acetyl phosphate Streptococcus pneumoniae naturally uses pyruvate oxidase to kill off competing bacteria when infecting the lungs. Having already demonstrated its activity and antibacterial properties in vivo, pyruvate oxidase was an attractive oxidase with which to work. Even though our activated engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough H2O2 to be cytotoxic and then fissle, killing only themselves and little else. To avoid this problem, an E. coli catalase will be expressed by default, and then turned off shortly after pyruvate oxidase expression is triggered. We are accomplishing this regulation by putting katG (on of two E. coli catalase genes) behind the TetR sensitive promoter (pTet) and having TetR co-transciptionally expressed with the oxidase. The time it takes for TetR to accumulate in the cell provides the delay in repressing KatG expression. To ensure KatG is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted. The final strain will have deletions of both catalases, ensuring that we have complete control over catalase expression. System DesignThe entire system will be engineered onto a high copy plasmid in E. coli as seen in Figure 3. Promoters are shown as bent, thin arrows, genes as thick arrows, ribosome binding sites as blue ovals, and a double transcriptional terminator as two red circles. Initially, the system will function independently, with luxR under a constitutive promoter. Later, luxR will be put under the control of a recombinase system that will only turn on the pathway randomly in a subset of cells. This will be one of three fates the master strain will be able to differentiate into. For more information, visit the population variation page. An ideal system would behave as outlined in Figure 4. The figure shows the relative abundances of the proteins and molecules used in the system. It is only meant to show if a particular species is relatively high or low, not its absolute concentration.
ResultsConstructsThe constructs shown in figure 5 were made using the standard assembly method. They were made to test the various regulatory elements and the functionality of the oxidase and catalase. Characterization of LuxR Inducer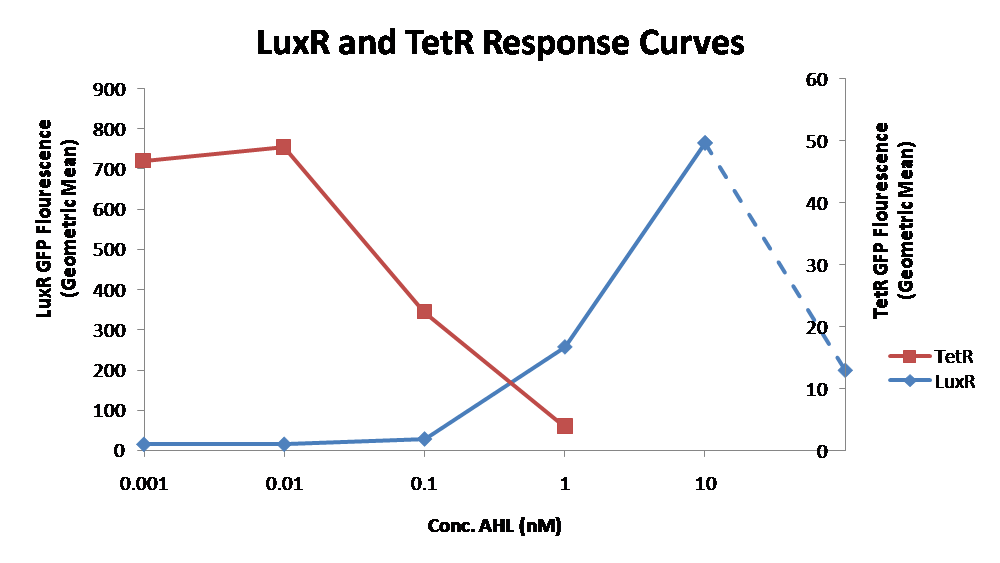 Figure 6 - Characterization of LuxR inducer and TetR inverter by flow cytometry. The figure shows the fluorescence of GFP in the constructs luxR-GFP and luxR-tetR-GFP. The left axis corresponds to the LuxR data and the right axis corresponds to the TetR data. At 100 nM AHL, cells with the luxR-GFP construct grew very poorly. The luxR inducer was characterized using the luxR-GFP construct. GFP provides a relative gauge of expression levels of SpxB in the final construct. The cultures were grown over a range of AHL concentrations and analyzed by flow cytometery. Figure 6 shows that the LuxR inducer behaves as expected. The system shows tight repression in the uninduced state, and begins to show significant expression at 1nM AHL. There is a 50 fold difference between the uninduced and fully induced (10nM AHL) state. The sudden drop at 100nM AHL is explained by the fact that these cells grew very poorly, possibly the result of toxic proteins loads. The LuxR inducer functions well if the concentration of AHL is kept below 10nM. Characterization of TetR InverterThe TetR inverter was characterized by using a luxR-tetR-GFP construct. In this context, GFP provides a relative gauge of expression levels of KatG in the final construct. Figure 6 shows that the tetracycline inverter behaved as expected. At high concentrations of AHL, the TetR inverter shows near full repression, as compared to the lower concentrations of AHL. Similar to the LuxR inducer, the TetR inverter showed a 50 fold difference in expression between the uninduced (low AHL) and induced (high AHL) states. However the absolute maximum level of expression from the TetR inverter was much lower than that from the LuxR inducer (compare the scale of the left TetR axis to the scale of the left LuxR axis). The low expression levels can be explained by the leakiness of the LuxR promoter, resulting in a basal level of repression from the TetR inverter. The low expression level is advantageous for the engineered E. coli because catalase is a very efficient scavenger of hydrogen peroxide. The low expression level ensures there is enough KatG to buffer any leaky pyruvate oxidase expression, yet still clear quickly from the cytoplasm. Additionally, in the uninduced state there should be extremely low levels of KatG. The LAA degradation tag should reduce KatG levels even further. Pyruvate Oxidase Produces Hydrogen Peroxide in vivo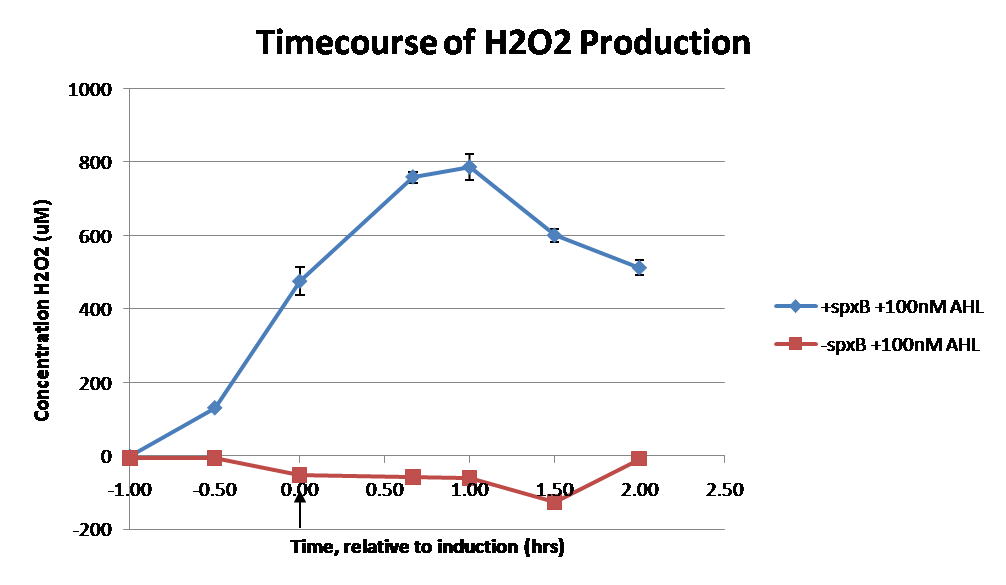 Figure 7: Timecourse of H2O2 production in JI377 cells with the luxR-spxB construct in SOC. JI377 cells were back diluted 1:100 and grown to an OD600 of ~0.8. Hydrogen peroxide in the supernatant was assayed every half hour. The arrow indicates induction with AHL at time 0 hr. Error bars represent one standard deviation(n=3) and are too small to be visible on the negative control. Using the H2O2 production assay protocol, JI377 (ΔahpCF'kan::'ahpFΔ(katG17::Tn10) katE12::Tn10) cells expressing pyruvate oxidase behind the LuxR inducer were able to produce significant amounts of hydrogen peroxide when cultured in rich media, reaching a maximum of 800 µM H2O2 in 2 hours (see figure 7).This is significant because the engineered cells can produce more than the minimum inhibitory concentration (MIC) for hydrogen peroxide of some human pathogens 2. The decline in hydrogen peroxide concentration after the peak is explained by the reaction of hydrogen peroxide with the SOC media, since H2O2 has a half life of about 3.5 hours in SOC (data not shown). The most surprising finding was that the cells began expressing hydrogen peroxide a full hour before they were induced with AHL. The current hypothesis is that some component of the rich SOC media mimicked AHL and activated LuxR. Uninduced expression from LuxR was also observed when culturing cell transformed with the luxR-GFP construct in LB media, as cell pellets were visibly green. However, no such autoinduction occurred in the defined minimal media M9. It would be interesting to see if LuxR spontaneously activates expression in a rich defined media, such as Neidhardt. However, the significant finding is that pyruvate oxidase is capable of producing bacteriostatic amounts of hydrogen peroxide in a short amount of time in vivo. Functionality of KatG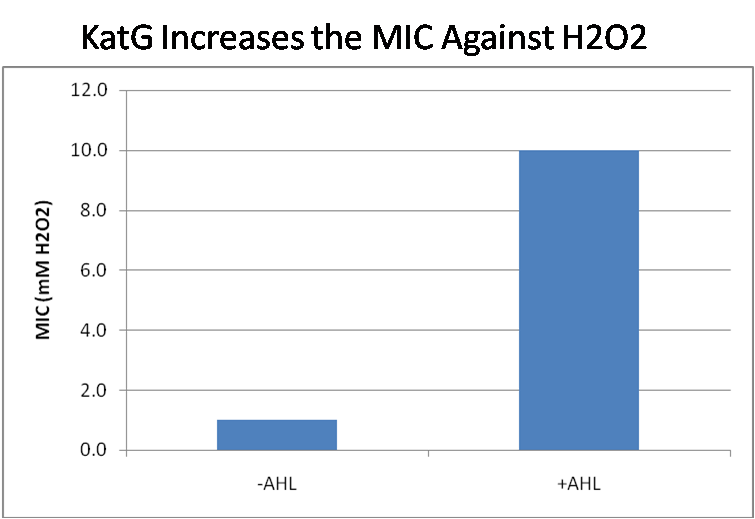 KatG +LAA confers hydrogen peroxide resistance. Saturated cultures of SN0037 cells with the construct luxR-katG were backdiluted 1:50 into SOC with or without AHL (0 or 100nM respectively) and various concentrations of hydrogen peroxide. The minimum inhibitory concentrations (MIC) for each condition are plotted. Functional expression of KatG from the luxR-katG construct protects cells from hydrogen peroxide. Figure 5 shows that expression of the KatG +LAA degradation tag in catalase deficient cells (SN0037, ΔkatGΔkatE) increases their measured minimum inhibitory concentration (MIC) by an order of magnitude. It is significant in that the MIC range of 3.0 - 10.0 mM H2O2 is well above to the MIC value of wild type E. coli (MIC = 0.8 uM). Even with a degradation tag, KatG is still functional and is sufficient to protect the cell from exogenous hydrogen peroxide. In an uninduced state, KatG will be able to buffer the engineered cell from leaky hydrogen peroxide production and should be degraded on the order of minutes because of the LAA tag. However in the final engineered strain, KatG will be expressed from behind the tetracycline inverter, not the LuxR system. Further tests will be necessary to ensure that the reduced expression levels from tetracycline inverter still produce enough KatG to raise the MIC of SN0037. The important result is that KatG +LAA is functionally expressed and is able to raise the MIC of SN0037. Coculture Assay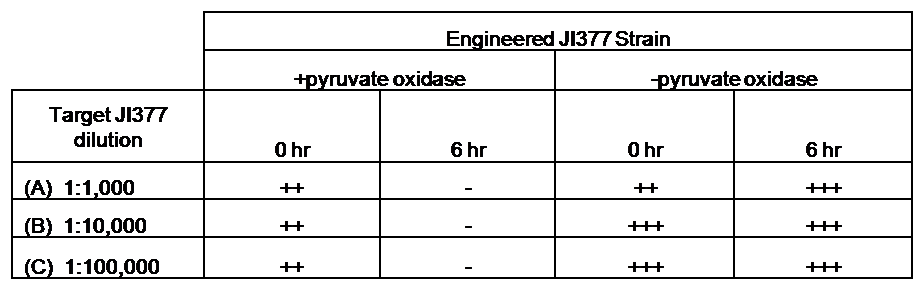 Table 1 - CFU counts of a competing E. coli strain before and after co-culturing with the engineered strain. Target JI377 strain (AmpR KanR) was inoculated in various amounts (A, B and C) into 2.5 mL SOC of engineered JI377 strain (AmpR). Production of hydrogen peroxide was induced with 10 nM AHL upon target strain inoculation. Cultures were serially diluted and plated onto LB+Kan at the given times. Key: +++ = lawn; ++ = many colonies, + = few colonies, - = no colonies. Finally, we wanted to see if the engineered JI377 cells could directly kill or inhibit the growth of a competing JI377 strain (due to time constraints, only JI377 with a luxR-spxB construct could be tested). We carried out co-culture assays in which varying amounts of the target strain were introduced to a heavy (OD600 of ~0.8) culture of the engineered strain, and incubated the co-culture for 6 hours after induction with AHL. Introducing the target strain to a dense population of our engineered E. coli was meant to mimic the situation in the gut where in invading pathogen will face an already established culture of engineered E. coli. Survival of the target strain was assayed by a colony forming unit (CFU) count both immediately upon introduction and after 6 hours of co-culturing. Table 1 summarizes the results. The CFU data show that the engineered strain was able to kill the target JI377 strain, while the target strain was unaffected or was able to grow in co-cultures with the negative control (JI377 with no pyruvate oxidase). ConclusionWe have successfully demonstrated the overproduction of hydrogen peroxide in E. coli (JI377) using the pyruvate oxidase from S. pneumoniae. This “oxidative burst” is sufficient to kill a competing strain of JI377 cells in co-culture assays. A genetic circuit was also constructed to regulate expression of pyruvate oxidase and catalase in response to the quorum sensing molecule acylhomoserine lactone. AHL and other quorum sensing molecules have been shown to be associated with the production of virulence factors of enteric pathogens, most notably Vibrio cholerae 1. Flow cytometry experiments showed that in a defined minimal media, the control circuit behaves as expected over a wide range of AHL concentrations. However in richer medias, the LuxR inducer is able to spontaneously turn on. The cause of this is still unknown, but it is possible some molecule in the undefined media closely mimics AHL. Overall, the data shows that engineered E. coli are capable of producing sufficient hydrogen peroxide to kill competing bacteria. References
|
 "
"