Team:University of Washington/Project
From 2008.igem.org
(→Construction of the Vector-Jector) |
|||
| (109 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
</html> | </html> | ||
| - | {| style="color:Gold;background-color:#500050;border | + | {| style="color:Gold;background-color:#500050;border: none" cellpadding="4" cellspacing="5" border="5" width="99%" align="center" |
| - | !align="center"; style="border- | + | !align="center";style="border: none;" |[[Image:WashingtonColorSeal-21-clip.gif|50px]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington|<font color="gold">Home</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Team|<font color="gold">The Team</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a inset 3px;" |[[Team:University_of_Washington/Project|<font color="gold">The Project</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Modeling|<font color="gold">Modeling</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Notebook|<font color="gold">Notebook</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Protocols|<font color="gold">Protocols</font>]] |
| - | !align="center"; style=" | + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Parts|<font color="gold">Parts Submitted <br>to the Registry</font>]] |
| - | !align="center"; style=" | + | !align="center"; style="background-color:#03943d;border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Measurement Kit|<font color="gold">Measurement Kit</font>]] |
| - | !align="center"; style="border | + | !align="center"; style="background-color:#c42317;border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/SeToB|<font color="gold">SeToB</font>]] |
| + | !align="center"; style="border: #6b0c6a outset 3px;" |[[Team:University_of_Washington/Safety|<font color="gold">Safety</font>]] | ||
|} | |} | ||
=Introduction: Controlled Trans-Kingdom Genetic Transfer= | =Introduction: Controlled Trans-Kingdom Genetic Transfer= | ||
| - | |||
| - | + | Bacteria have several mechanisms for transferring genes between individuals, and even between species, that is independent of traditional heritable genetics. Bacterial conjugation is one such mechanism in which genetic information is transferred horizontally. During conjugation, a bacterial cell constructs a sex pilus nanotube that connects donor and recipient bacteria. A conjugative plasmid directs the construction of the structural proteins and enzymes that bind to a specific DNA sequence (called an "origin of transfer" or OriT) to direct replication and transfer of the conjugative plasmid. In addition, any other plasmids that contain an appropriate OriT will be transferred as well. In this manner, recipient bacteria can accept and express genes separate from their chromosomal DNA. These genes may encode novel functions such as metabolic abilities or resistance to antibiotics. This mode of genetic transfer is fascinating on many levels, including its possible role in evolution and its potential use for genetic alteration. | |
| - | |||
| - | + | In addition to horizontal gene transfer between bacteria, it has been demonstrated that bacteria can also transfer genetic material, via conjugation, between phylogenetic kingdoms [1]. Escherichia coli, a common laboratory bacterium, is capable of transferring shuttle vectors that contain an appropriate OriT to the eukaryote Saccharomyces cerevisiae, more commonly known as baker's yeast. If a yeast origin of replication is present on the shuttle plasmid, S. cerevisiae is able to maintain the plasmid and express the acquired genes. This phenomenon is remarkable considering the vast evolutionary distance between these organisms [1]. The goal of our project is to control this process of trans-kingdom genetic transfer by engineering genetic devices that induce genetic transfer under user-defined conditions. This genetically engineered machine could see future use in gene therapy or for eukaryotic transformation. | |
| - | = | + | =Construction of the Vector-Jector= |
| - | + | In order to construct this controlled trans-kingdom genetic transfer system (which we named the Vector-Jector), we created a design that implements the following modules (Figure 1): a yeast Signaling Module that allows S. cerevisiae to signal for genetic transfer (#1 in Figure 1); an Activity Decision Module that detects environmental signals and uses this information to generate an output (in the form of PoPS) (#2 in Figure 1); a Conjugative Machinery Control Module that connects PoPS to production of the conjugative apparatus (#3 in Figure 1); and a Conferred Ability Module that bestows some novel bioloigcal function to S. cerevisiae (#4 in Figure 1). | |
| - | |||
| - | + | [[Image:uwbigpicture.jpg|400px|center]] | |
| + | '''FIGURE 1 - General Architecture of the Vector-Jector''' | ||
| - | |||
| - | + | Our modular design for trans-kingdom genetic transfer could be adapted for use with various organisms and under different circumstances. To demonstrate the proof-of-principle for our device, we chose to implement the following scenario: when E. coli and S. cerevisiae are in close proximity and lactose is the primary metabolic source, E. coli cells will construct the conjugative apparatus and transfer the yeast shuttle vector. This yeast shuttle vector confers the ability to digest lactose upon S. cerevisiae, an ability it does not have naturally. The Vector-Jector only carries out genetic transfer under specific environmental conditions. This scenario is a test case for accomplishing gene transfer under user-defined conditions. | |
| - | + | ||
| - | |||
| - | + | How we implemented the circuitry is depicted in Figure 2. | |
| - | + | *Module 1, the Signaling Module, exists as a means for the metabolically stressed yeast to signal for transfer of the conjugative plasmid. To create this, we planned to construct a yeast expression plasmid containing the gene to produce AHL under the control of a native yeast promoter that is activated in the presence of lactose and the absence of glucose. Repressing the system in the presence of glucose is necessary since S. cerevisiae preferentially metabolizes glucose, so if glucose is present in the environment, gene transfer from the Vector-Jector is not required for S. cerevisiae growth regardless of the presence of lactose. | |
| + | *Module 2, the Activity Decision Module, represents the mechanism by which E. coli detects user-defined environmental signals and generates a PoPS output when the necessary conditions are met. Our specific conditions are the presence of lactose, the absence of glucose, and the presence of a signal from the metabolically stressed S. cerevisiae. Therefore, we required circuitry that provided a PoPS output when these three conditions are met. Using standard BioBrick assembly methods, we placed the gene for LuxR under the control of the full-length, wild type lac promoter, which is activated in the presence of lactose, and repressed in the presence of glucose. LuxR can then activate gene expression from the pLux promoter, but only if AHL is present. In this way, all three conditions must be met to generate a PoPS output. To demonstrate the modularity of the Vector-Jector, we also constructed an Alternative Activity Decision Module, which generates a PoPS input with a different inputs - in this case, arabinose, anhydrotetracycline, and AHL. | ||
| + | |||
| + | *Module 3, the Conjugation Machinery Control Module, connected the PoPS input from Module 2 to generation of the conjugative apparatus. To do this, we selected genes that encode important regulatory and structural proteins and regulated their expression using pLux (instead of their native promoters). | ||
| + | |||
| + | *Module 4 (Not shown in Figure 2), the Conferred Ability Module, needed to transfer to S. cerevisiae the ability to metabolize lactose. To do this, we used a previously-designed shuttle plasmid, capable of maintenance and replication in both bacteria and yeast and containing the correct OriT [5], and inserted the gene for beta-galactosidase, the enzyme that metabolizes lactose, which is functional in S. cerevisiae. The conjugative apparatus produced by the Conjugation Machinery Control Module would recognize this OriT and direct its transfer into S. cerevisiae. | ||
| + | |||
| + | [[Image:uwsuboverview.gif|850px|center]] | ||
| + | '''FIGURE 2 - Implementation of Modules 1-3 of the Vector Jector''' | ||
| + | |||
| + | |||
| + | We also generated a software tool called SeToB, which would make the design of projects like ours easier using information from the Registry of Standard Biological Parts (see SeToB tab). | ||
| + | |||
| + | =Individual Projects= | ||
| + | |||
| + | ==Module #1: Signaling Module <font size="2"><b>- Biosynthesis of the AHL distress signal (in Yeast)</b></font>== | ||
| + | |||
| + | [[Image:uwAHL.gif|150px|left]] | ||
| + | Ultimately, our goal is to place bacterial conjugation under the control of an external distress signal from a metabolically-stressed yeast cell. Thus, it is necessary to engineer ''S. cerevisiae'' to produce the AHL signaling molecule. Our proposed method to accomplish this is to clone the AI synthase gene (BBa_C0161) into a nonstandard yeast expression plasmid, which should theoretically result in the conversion of amino acid precursors into the AHL molecule. To test whether the yeast transformants are indeed capable of AHL signaling, they can be tested in co-culture with the AHL receiver cells (BBa_T9002). Although AHL production will initially be under control of a constitutive promoter, eventually we may place it under control of a glucose-repressible promoter such as pJEN1 [2], so that the yeast will produce AHL proportionally in response to glucose starvation. | ||
| + | |||
| + | ==Module #2: Activity Decision Module <font size="2"><b>- LuxR production under pLac/pAraC+TetR in bacteria</b></font>== | ||
| + | |||
| + | ===Regulators: pLac=== | ||
| + | [[Image:AND_LUX.gif|200px|left]] | ||
| + | Lactose is a disaccharide composed of glucose and galactose, and in order for an organism to metabolize lactose, it first must be broken-up into these constituent monomers. β-galactosidase is an enzyme capable of doing just that, and is naturally produced by ''E. coli''. However, ''E. coli'' cleverly regulate their production of this enzyme to times only when it’s required. Being that lactose is a less efficient fuel than simple glucose, ''E. coli'' only want to metabolize lactose when glucose is absent. This regulation is carried out at the transcriptional level by putting the β-galactosidase gene (and 2 other genes) under the control of a promoter (the lac promoter) which is inhibited by LacI and induced by CAP. LacI is a molecule that is constitutively produced by ''E. coli''; it can either bind to the promoter or bind to lactose, meaning that LacI only inhibits the promoter in the absence of lactose. CAP is another molecule naturally present in ''E. coli'' which is only free to bind to the lac promoter in the absence of glucose. Given that gene repression is stronger than activation, the production of β-galactosidase is only carried out when required. | ||
| + | |||
| + | In order to have LuxR produced only when glucose is absent and lactose is present, the LuxR protein generator BioBrick (part BBa_I0462) was placed under the control of the lac promoter (part BBa_R0010). The lac promoter is a commonly used transcriptional regulator, as it enables genes to be turned on/off with addition/subtraction of lactose (or IPTG) from the growth medium. On the other hand, the use of glucose as a means to regulate the activity of the promoter is not common. Thus, promoter part BBa_R0010 had to be tested to verify that it indeed behaves as the natural lac promoter does (i.e. IPTG can only successfully activate the promoter in the absence of glucose). | ||
| + | |||
| + | To carry out such a test, part BBa_R0010 was placed in front of the GFP generator BioBrick (part BBa_E0240). An assay was then carried out that characterized the promoter activity in varying concentrations of IPTG and glucose: 25 wells of a 96-well plate were filled with the mid-log culture of a LacI expressing ''E. coli'' strain (DH5a+LacIq) grown in M9 minimal media. IPTG and glucose was then added to the wells in different amounts. The GFP fluorescence of each well was then measured over a six hour period. As the plot shows below, GFP has the highest expression in the well with the highest concentration of IPTG and no glucose, and the lowest expression in the well with the highest glucose concentration and no IPTG. Additionally, in wells with both glucose and IPTG, the repression of the glucose proved stronger than the activation of the IPTG. Thus the promoter part behaves just as required. After parts BBa_R0010 and BBa_I0462 were ligated together, they were placed inside the low/medium copy, kan resistant plasmid pSB3K3. This new composite part was given name BBa_K109702. | ||
| + | |||
| + | {|style="background-color:#FFdd00" border="0" width="800px" align=center | ||
| + | | | ||
[[Image:IGEMdataPlot2.jpg|center|800px]] | [[Image:IGEMdataPlot2.jpg|center|800px]] | ||
| + | |- | ||
| + | |'''Activity of pLac in Varying Concentrations of IPTG and Glucose'''<br\> | ||
| + | <p><small>A GFP generator gene (part BBa_E0240) was placed under the control of the Lac promoter (part BBa_R0010). Fluorescence was then measured under varying concentrations of IPTG and glucose to characterize the promoter's activity.</small></p> | ||
| + | |} | ||
| - | ===Regulators: | + | ===Regulators: pAraC+TetR=== |
| + | {| style="background-color:#FFdd00" border="0" | ||
| + | | | ||
| + | [[Image:AND_AT.gif|200px|left]] | ||
| + | Since lactose and glucose play an important role in regular cellular systems, there may be some complication that could affect our genetic circuits or how cells function in general. To try avoiding problems, we make alternative promoter with AraC and TetR binding sites. AraC is an activator induced by arabinose sugar. TetR is a repressor that is made inactive by tetracycline(tc) or anhydrotetracycline(aTc). Both arabinose and tc/aTc are less essential to cells under standard lab conditions, to control LuxR production. | ||
| - | + | In the research "Programming gene expression with combinatorial promoters" [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html <small>link</small>] by Robert Sidney Cox, III, Michael G Surette, and Michael B Elowitz [3], it was shown that having araC at distal region(upstream -35) and tetR at proximal region(downstream -10) made the most AND-gate promoter among the combination of AraC and TetR hybrid regulators. | |
| - | + | The first approach to make the promoter is to insert TetR binding sites into existing AraC promoter BioBrick (BBa_R0080) by [[QuikChange_Mutagenesis|QuikChange Mutagenesis method]]. This approach did not work out. Some possible reasons were that the primers were not well designed (i.e. homology of primers and parental plasmid were not long enough for primers to bind to parental plasmid.), and the materials were not correct or effective(reactions were made from existing lab's material without a kit). | |
| - | The | + | The second approach is to PCR amplify the promoter region containing araC and tetR from the clone D29 received from Elowitz's lab. Using primers with BioBrick prefix and suffix hangings, we succeed making a BioBrick of the promoter part(BBa_K109200), tested it with GFP reporter(BBa_E0240)-shown below-, and putting it in front of LuxR gene(BBa_I0462). |
| + | | | ||
| + | [[Image:AraC+TetRmini.jpg]] | ||
| + | |} | ||
| + | [[Image:uwpATGFPassay.jpg|900px|center]] | ||
| - | The | + | The promoter-GFP plasmid was transformed into ''E.coli'' strain MG1655Z1[3] that constitutively produces TetR and AraC protein. The culture was grown in LB and put into arabinose sugar and aTc that were already mixed in 5x5 wells of 96-well plate. The OD and fluorescence were measured using 96-well plate reader every 10 minutes for 18 hours. Even though the overall fluorescence data is low, the normalized(fluorescence/OD) data implies the AND logic. From the graph above, there is a trend of increasing GFP expression as aTc and arabinose become more concentrated. However, there was a spike at the edge of the graph, especially when arabinose is zero and aTc is low. This might have resulted from wet-lab errors that contaminated the mixtures when trying to put the film (to prevent evaporation) onto the plate. |
| - | + | ==Module #3: Conjugation Machinery Control <font size="2"><b>- Replacement of Conjugative Promoters with an Inducible Promoter</b></font>== | |
| - | + | [[Image:AND_CON.gif|300px|right]] | |
| - | + | The next step is to place conjugative genes under control of pLuxR (BBa_R6002), an AHL inducible promoter. The iGEM team used two approaches to place these genes under control of pLuxR. | |
| - | The | + | The first approach was to augment the conjugative plasmid using Lambda Red recombineering with the intent of replacing native promoters with pLuxR. We identified two target promoters that are crucial control points in conjugation. The first target, pTrbA, encodes a transcriptional regulator for the TraII conjugation operon. The other target, pKorA, expresses a global regulatory protein with multiple binding sites across the conjugative plasmid's genome. The KorA regulatory protein is an attractive target because it controls a genetic OR switch that represses vegetative replication while promoting conjugation--a fascinating genetic component that helps the plasmid balance the metabolic demand it places on the host cell. [4] |
| - | + | The second approach was to knockout essential conjugation genes and complement them on secondary plasmids under control of the AHL inducible promoter. Again, the korA and trbA genes were targeted. We also targeted the trbC gene, which encodes the pilin protein from which the mating pilus is constructed. Using this approach, we were able to construct standardized BioBricks--but with the drawback that we would now have to maintain this additional complementary plasmid in our final system. | |
| - | [[Image:RP4 conjugation.gif| | + | {|style="background-color:#FFdd00" border="0" cellpadding="2" |
| - | + | |width=45% align=center|[[Image:RP4 conjugation.gif|400px]] | |
| - | < | + | '''Illustration of Tra1 Conjugation Machinery'''<br\> |
| + | <p><small>Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [http://www.ncbi.nlm.nih.gov/pubmed/11976307 link].</small></p> | ||
| + | |[[Image:uwlamdared.gif|550px]] | ||
| + | |} | ||
| + | |||
| + | ==Demonstration of ''E. coli - S. cerevisiae'' Genetic Transfer== | ||
| + | |||
| + | Before we could control ''E. coli - S. cerevisiae'' genetic transfer, we had to first develop a protocol and demonstrate (uncontrolled) conjugative transfer between these two organisms, shown below. | ||
| + | Shown are four separate experiments. In each experiment, bacteria was mixed with yeast, incubated to allow for conjugative transfer, and plated on yeast media that contains antibacterial antibiotics and lacks the essential amino acid leucine. Thus, any growth on these plates is yeast that can generate leucine. The bacteria contained a yeast shuttle plasmid with an OriT that conferred upon yeast the ability to generate leucine (right half of plates). We know that this genetic transfer occurred by conjugation since bacteria lacking the conjugative plasmid were unable to transfer the shuttle plasmid (left half of plates). | ||
| - | <table width="98%" align="center" border="1"> | + | <table style="background-color:#FFdd00" width="98%" align="center" border="1"> |
<tr><th align="left" width="28%">''Experiment #''</th><th>1</th><th>2</th><th>3</th><th>4</th></tr> | <tr><th align="left" width="28%">''Experiment #''</th><th>1</th><th>2</th><th>3</th><th>4</th></tr> | ||
<tr><th align="left">Bacteria (OD600@0.15 ≈ 3E7 bac/mL)</th><td>250uL</td><td>200uL</td><td>100uL</td><td>50uL</td></tr> | <tr><th align="left">Bacteria (OD600@0.15 ≈ 3E7 bac/mL)</th><td>250uL</td><td>200uL</td><td>100uL</td><td>50uL</td></tr> | ||
| Line 106: | Line 153: | ||
|} | |} | ||
| - | == Yeast Shuttle Vector == | + | ==Module #4: Conferred Ability Module<font size="2"><b>- Yeast Shuttle Vector</b></font>== |
| - | + | ||
| - | + | [[Image:yst.jpg|right|400px]] | |
| - | + | Another important part in our controlled trans-kingdom conjugation project is the yeast shuttle vector, which confers a novel ability to yeast. This plasmid is stored and maintained in ''E. coli'', transferred to yeast via conjugation, then stored and maintained in yeast. Therefore, this yeast shuttle plasmid requires a bacterial origin of replication, a stable yeast origin of replication, a conjugative origin of transfer for RP4 mobilization, a bacterial selectable marker that confers ampicilin resistance, and a gene which imparts the ability to synthesize leucine. A plasmid containing each of these features, pAC88, was obtained from the University of Leicester where it was designed for prior research [5]. | |
| - | + | ||
| + | The pAC88 yeast shuttle vector, while suitable for our experiment, does not adhere to the standard BioBricks format. It contains many extraneous restriction sites strewn around the plasmid and within part sequences, while also lacking BioBrick restriction sites which flank the part sequences. Therefore, pAC88 cannot be used in standard assembly or to extract parts. In an effort to provide the synthetic biology community with a more useful, standardized yeast vector, the necessary feature for maintainance, selection, and transfer are being incorporated into the BioBrick construction base vector. | ||
| + | Insertion of an adaptive gene into pAC88 was also attempted. Kluyveromyces lactis, a species of milk-yeast, posesses a region of chromosomal DNA important in the production lactose digestion machinery. LAC4 and LAC12, two genes that encode for yeast galactosidase and lactose permease, respectively, are located near each other on the chromosome, and are divergently transcribed. This LAC region was amplified and inserted into an XhoI restriction site on pAC88. However, yeast transformants remained unable to digest lactose. | ||
| + | ---- | ||
[1] Sprague, George F., and Jack A. Heinemann. “Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast.” Nature. Institute of Molecular Biology. University of Oregon. 340. 20 Jul 1989. | [1] Sprague, George F., and Jack A. Heinemann. “Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast.” Nature. Institute of Molecular Biology. University of Oregon. 340. 20 Jul 1989. | ||
| - | [2] Cashmore, Annette M., Steven Bates, and Brian M. Wilkins. “IncP Plasmids Are Unusually Effective in Mediating Conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 Mating System.” Journal of Bacteriology. Dec 1998. pp 6538-6543. | + | [2] Chambers P, Issaka A, Palecek SP. "Saccharomyces cerevisiae JEN1 promoter activity is inversely related to concentration of repressing sugar." Appl Environ Microbiol. 2004 Jan;70(1):8-17. |
| + | |||
| + | [3] Robert Sidney Cox, III, Michael G Surette, and Michael B Elowitz. "Programming gene expression with combinatorial promoters." Nature. Institute of Molecular Biology. 145. 13 November 2007 | ||
| + | |||
| + | [4] Zatyka M, Jagura-Burdzy G, Thomas CM. "Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids." J Bacteriol. 1997 Dec;179(23):7201-9. | ||
| + | |||
| + | [5] Cashmore, Annette M., Steven Bates, and Brian M. Wilkins. “IncP Plasmids Are Unusually Effective in Mediating Conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 Mating System.” Journal of Bacteriology. Dec 1998. pp 6538-6543. | ||
| - | |||
<!--- The Mission ---> | <!--- The Mission ---> | ||
<!---== Your abstract ==---> | <!---== Your abstract ==---> | ||
Latest revision as of 04:40, 30 October 2008

| Home | The Team | The Project | Modeling | Notebook | Protocols | Parts Submitted to the Registry | Measurement Kit | SeToB | Safety |
|---|
Introduction: Controlled Trans-Kingdom Genetic Transfer
Bacteria have several mechanisms for transferring genes between individuals, and even between species, that is independent of traditional heritable genetics. Bacterial conjugation is one such mechanism in which genetic information is transferred horizontally. During conjugation, a bacterial cell constructs a sex pilus nanotube that connects donor and recipient bacteria. A conjugative plasmid directs the construction of the structural proteins and enzymes that bind to a specific DNA sequence (called an "origin of transfer" or OriT) to direct replication and transfer of the conjugative plasmid. In addition, any other plasmids that contain an appropriate OriT will be transferred as well. In this manner, recipient bacteria can accept and express genes separate from their chromosomal DNA. These genes may encode novel functions such as metabolic abilities or resistance to antibiotics. This mode of genetic transfer is fascinating on many levels, including its possible role in evolution and its potential use for genetic alteration.
In addition to horizontal gene transfer between bacteria, it has been demonstrated that bacteria can also transfer genetic material, via conjugation, between phylogenetic kingdoms [1]. Escherichia coli, a common laboratory bacterium, is capable of transferring shuttle vectors that contain an appropriate OriT to the eukaryote Saccharomyces cerevisiae, more commonly known as baker's yeast. If a yeast origin of replication is present on the shuttle plasmid, S. cerevisiae is able to maintain the plasmid and express the acquired genes. This phenomenon is remarkable considering the vast evolutionary distance between these organisms [1]. The goal of our project is to control this process of trans-kingdom genetic transfer by engineering genetic devices that induce genetic transfer under user-defined conditions. This genetically engineered machine could see future use in gene therapy or for eukaryotic transformation.
Construction of the Vector-Jector
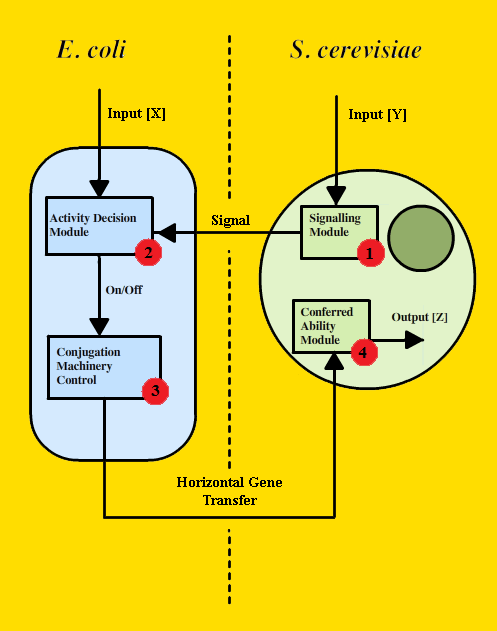
In order to construct this controlled trans-kingdom genetic transfer system (which we named the Vector-Jector), we created a design that implements the following modules (Figure 1): a yeast Signaling Module that allows S. cerevisiae to signal for genetic transfer (#1 in Figure 1); an Activity Decision Module that detects environmental signals and uses this information to generate an output (in the form of PoPS) (#2 in Figure 1); a Conjugative Machinery Control Module that connects PoPS to production of the conjugative apparatus (#3 in Figure 1); and a Conferred Ability Module that bestows some novel bioloigcal function to S. cerevisiae (#4 in Figure 1).
FIGURE 1 - General Architecture of the Vector-Jector
Our modular design for trans-kingdom genetic transfer could be adapted for use with various organisms and under different circumstances. To demonstrate the proof-of-principle for our device, we chose to implement the following scenario: when E. coli and S. cerevisiae are in close proximity and lactose is the primary metabolic source, E. coli cells will construct the conjugative apparatus and transfer the yeast shuttle vector. This yeast shuttle vector confers the ability to digest lactose upon S. cerevisiae, an ability it does not have naturally. The Vector-Jector only carries out genetic transfer under specific environmental conditions. This scenario is a test case for accomplishing gene transfer under user-defined conditions.
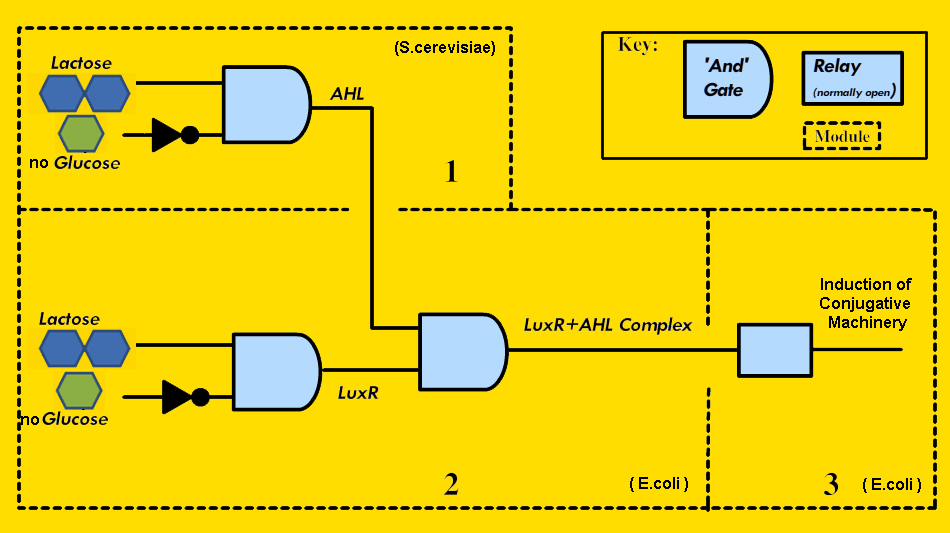
How we implemented the circuitry is depicted in Figure 2.
- Module 1, the Signaling Module, exists as a means for the metabolically stressed yeast to signal for transfer of the conjugative plasmid. To create this, we planned to construct a yeast expression plasmid containing the gene to produce AHL under the control of a native yeast promoter that is activated in the presence of lactose and the absence of glucose. Repressing the system in the presence of glucose is necessary since S. cerevisiae preferentially metabolizes glucose, so if glucose is present in the environment, gene transfer from the Vector-Jector is not required for S. cerevisiae growth regardless of the presence of lactose.
- Module 2, the Activity Decision Module, represents the mechanism by which E. coli detects user-defined environmental signals and generates a PoPS output when the necessary conditions are met. Our specific conditions are the presence of lactose, the absence of glucose, and the presence of a signal from the metabolically stressed S. cerevisiae. Therefore, we required circuitry that provided a PoPS output when these three conditions are met. Using standard BioBrick assembly methods, we placed the gene for LuxR under the control of the full-length, wild type lac promoter, which is activated in the presence of lactose, and repressed in the presence of glucose. LuxR can then activate gene expression from the pLux promoter, but only if AHL is present. In this way, all three conditions must be met to generate a PoPS output. To demonstrate the modularity of the Vector-Jector, we also constructed an Alternative Activity Decision Module, which generates a PoPS input with a different inputs - in this case, arabinose, anhydrotetracycline, and AHL.
- Module 3, the Conjugation Machinery Control Module, connected the PoPS input from Module 2 to generation of the conjugative apparatus. To do this, we selected genes that encode important regulatory and structural proteins and regulated their expression using pLux (instead of their native promoters).
- Module 4 (Not shown in Figure 2), the Conferred Ability Module, needed to transfer to S. cerevisiae the ability to metabolize lactose. To do this, we used a previously-designed shuttle plasmid, capable of maintenance and replication in both bacteria and yeast and containing the correct OriT [5], and inserted the gene for beta-galactosidase, the enzyme that metabolizes lactose, which is functional in S. cerevisiae. The conjugative apparatus produced by the Conjugation Machinery Control Module would recognize this OriT and direct its transfer into S. cerevisiae.
FIGURE 2 - Implementation of Modules 1-3 of the Vector Jector
We also generated a software tool called SeToB, which would make the design of projects like ours easier using information from the Registry of Standard Biological Parts (see SeToB tab).
Individual Projects
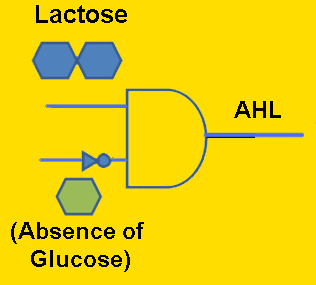
Module #1: Signaling Module - Biosynthesis of the AHL distress signal (in Yeast)
Ultimately, our goal is to place bacterial conjugation under the control of an external distress signal from a metabolically-stressed yeast cell. Thus, it is necessary to engineer S. cerevisiae to produce the AHL signaling molecule. Our proposed method to accomplish this is to clone the AI synthase gene (BBa_C0161) into a nonstandard yeast expression plasmid, which should theoretically result in the conversion of amino acid precursors into the AHL molecule. To test whether the yeast transformants are indeed capable of AHL signaling, they can be tested in co-culture with the AHL receiver cells (BBa_T9002). Although AHL production will initially be under control of a constitutive promoter, eventually we may place it under control of a glucose-repressible promoter such as pJEN1 [2], so that the yeast will produce AHL proportionally in response to glucose starvation.
Module #2: Activity Decision Module - LuxR production under pLac/pAraC+TetR in bacteria
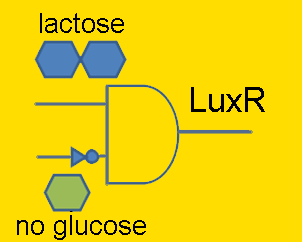
Regulators: pLac
Lactose is a disaccharide composed of glucose and galactose, and in order for an organism to metabolize lactose, it first must be broken-up into these constituent monomers. β-galactosidase is an enzyme capable of doing just that, and is naturally produced by E. coli. However, E. coli cleverly regulate their production of this enzyme to times only when it’s required. Being that lactose is a less efficient fuel than simple glucose, E. coli only want to metabolize lactose when glucose is absent. This regulation is carried out at the transcriptional level by putting the β-galactosidase gene (and 2 other genes) under the control of a promoter (the lac promoter) which is inhibited by LacI and induced by CAP. LacI is a molecule that is constitutively produced by E. coli; it can either bind to the promoter or bind to lactose, meaning that LacI only inhibits the promoter in the absence of lactose. CAP is another molecule naturally present in E. coli which is only free to bind to the lac promoter in the absence of glucose. Given that gene repression is stronger than activation, the production of β-galactosidase is only carried out when required.
In order to have LuxR produced only when glucose is absent and lactose is present, the LuxR protein generator BioBrick (part BBa_I0462) was placed under the control of the lac promoter (part BBa_R0010). The lac promoter is a commonly used transcriptional regulator, as it enables genes to be turned on/off with addition/subtraction of lactose (or IPTG) from the growth medium. On the other hand, the use of glucose as a means to regulate the activity of the promoter is not common. Thus, promoter part BBa_R0010 had to be tested to verify that it indeed behaves as the natural lac promoter does (i.e. IPTG can only successfully activate the promoter in the absence of glucose).
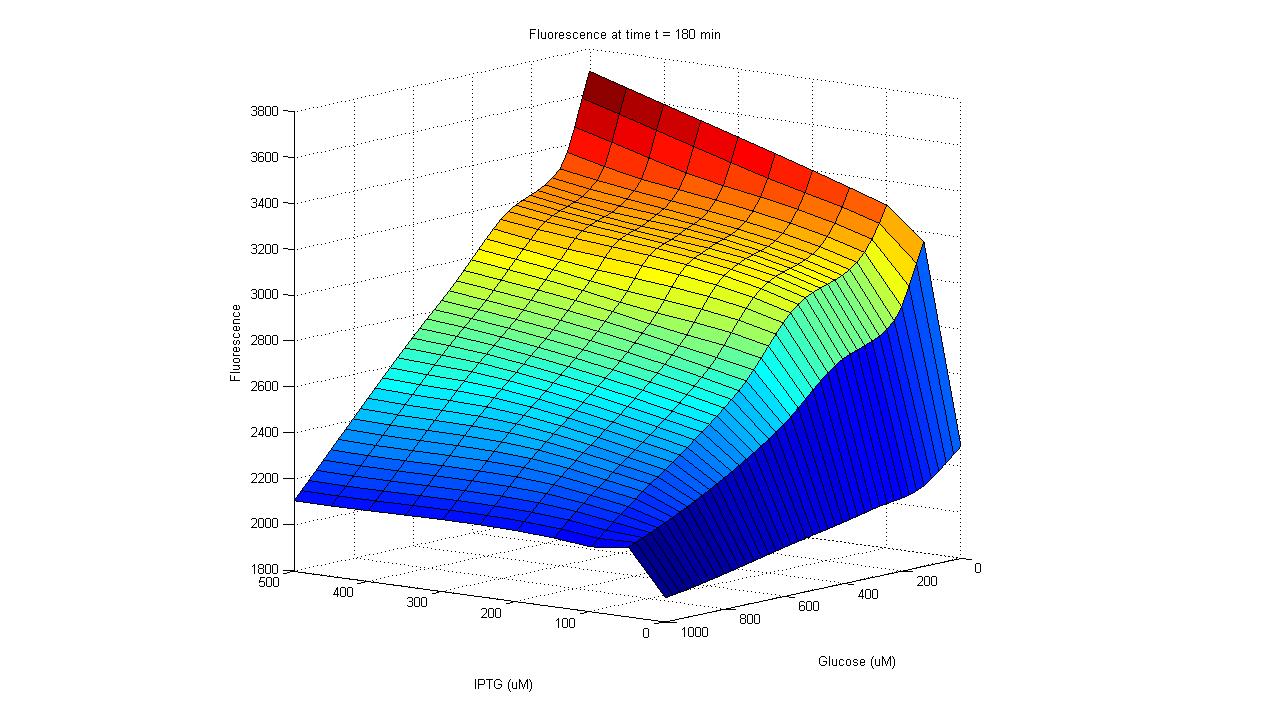
To carry out such a test, part BBa_R0010 was placed in front of the GFP generator BioBrick (part BBa_E0240). An assay was then carried out that characterized the promoter activity in varying concentrations of IPTG and glucose: 25 wells of a 96-well plate were filled with the mid-log culture of a LacI expressing E. coli strain (DH5a+LacIq) grown in M9 minimal media. IPTG and glucose was then added to the wells in different amounts. The GFP fluorescence of each well was then measured over a six hour period. As the plot shows below, GFP has the highest expression in the well with the highest concentration of IPTG and no glucose, and the lowest expression in the well with the highest glucose concentration and no IPTG. Additionally, in wells with both glucose and IPTG, the repression of the glucose proved stronger than the activation of the IPTG. Thus the promoter part behaves just as required. After parts BBa_R0010 and BBa_I0462 were ligated together, they were placed inside the low/medium copy, kan resistant plasmid pSB3K3. This new composite part was given name BBa_K109702.
| Activity of pLac in Varying Concentrations of IPTG and Glucose A GFP generator gene (part BBa_E0240) was placed under the control of the Lac promoter (part BBa_R0010). Fluorescence was then measured under varying concentrations of IPTG and glucose to characterize the promoter's activity. |
Regulators: pAraC+TetR
|
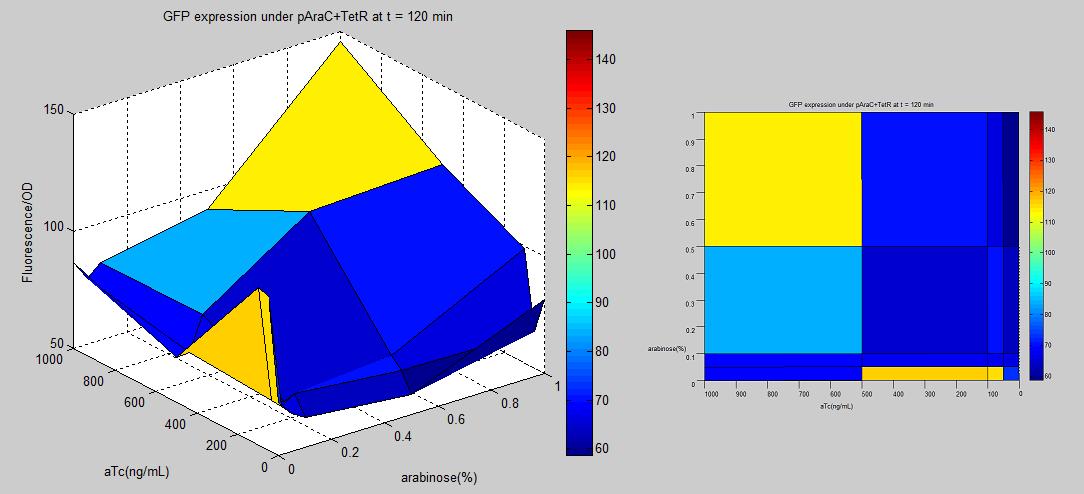
Since lactose and glucose play an important role in regular cellular systems, there may be some complication that could affect our genetic circuits or how cells function in general. To try avoiding problems, we make alternative promoter with AraC and TetR binding sites. AraC is an activator induced by arabinose sugar. TetR is a repressor that is made inactive by tetracycline(tc) or anhydrotetracycline(aTc). Both arabinose and tc/aTc are less essential to cells under standard lab conditions, to control LuxR production. In the research "Programming gene expression with combinatorial promoters" [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html link] by Robert Sidney Cox, III, Michael G Surette, and Michael B Elowitz [3], it was shown that having araC at distal region(upstream -35) and tetR at proximal region(downstream -10) made the most AND-gate promoter among the combination of AraC and TetR hybrid regulators. The first approach to make the promoter is to insert TetR binding sites into existing AraC promoter BioBrick (BBa_R0080) by QuikChange Mutagenesis method. This approach did not work out. Some possible reasons were that the primers were not well designed (i.e. homology of primers and parental plasmid were not long enough for primers to bind to parental plasmid.), and the materials were not correct or effective(reactions were made from existing lab's material without a kit). The second approach is to PCR amplify the promoter region containing araC and tetR from the clone D29 received from Elowitz's lab. Using primers with BioBrick prefix and suffix hangings, we succeed making a BioBrick of the promoter part(BBa_K109200), tested it with GFP reporter(BBa_E0240)-shown below-, and putting it in front of LuxR gene(BBa_I0462). |
The promoter-GFP plasmid was transformed into E.coli strain MG1655Z1[3] that constitutively produces TetR and AraC protein. The culture was grown in LB and put into arabinose sugar and aTc that were already mixed in 5x5 wells of 96-well plate. The OD and fluorescence were measured using 96-well plate reader every 10 minutes for 18 hours. Even though the overall fluorescence data is low, the normalized(fluorescence/OD) data implies the AND logic. From the graph above, there is a trend of increasing GFP expression as aTc and arabinose become more concentrated. However, there was a spike at the edge of the graph, especially when arabinose is zero and aTc is low. This might have resulted from wet-lab errors that contaminated the mixtures when trying to put the film (to prevent evaporation) onto the plate.
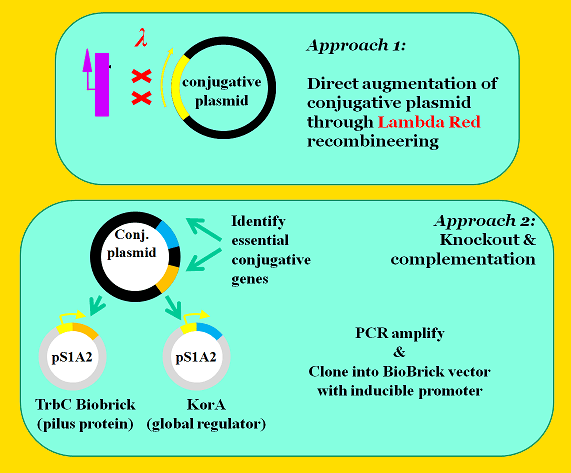
Module #3: Conjugation Machinery Control - Replacement of Conjugative Promoters with an Inducible Promoter
The next step is to place conjugative genes under control of pLuxR (BBa_R6002), an AHL inducible promoter. The iGEM team used two approaches to place these genes under control of pLuxR.
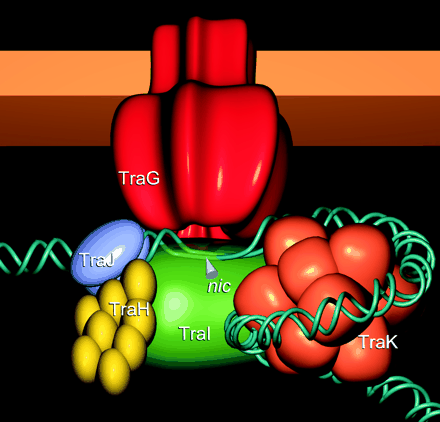
The first approach was to augment the conjugative plasmid using Lambda Red recombineering with the intent of replacing native promoters with pLuxR. We identified two target promoters that are crucial control points in conjugation. The first target, pTrbA, encodes a transcriptional regulator for the TraII conjugation operon. The other target, pKorA, expresses a global regulatory protein with multiple binding sites across the conjugative plasmid's genome. The KorA regulatory protein is an attractive target because it controls a genetic OR switch that represses vegetative replication while promoting conjugation--a fascinating genetic component that helps the plasmid balance the metabolic demand it places on the host cell. [4]
The second approach was to knockout essential conjugation genes and complement them on secondary plasmids under control of the AHL inducible promoter. Again, the korA and trbA genes were targeted. We also targeted the trbC gene, which encodes the pilin protein from which the mating pilus is constructed. Using this approach, we were able to construct standardized BioBricks--but with the drawback that we would now have to maintain this additional complementary plasmid in our final system.
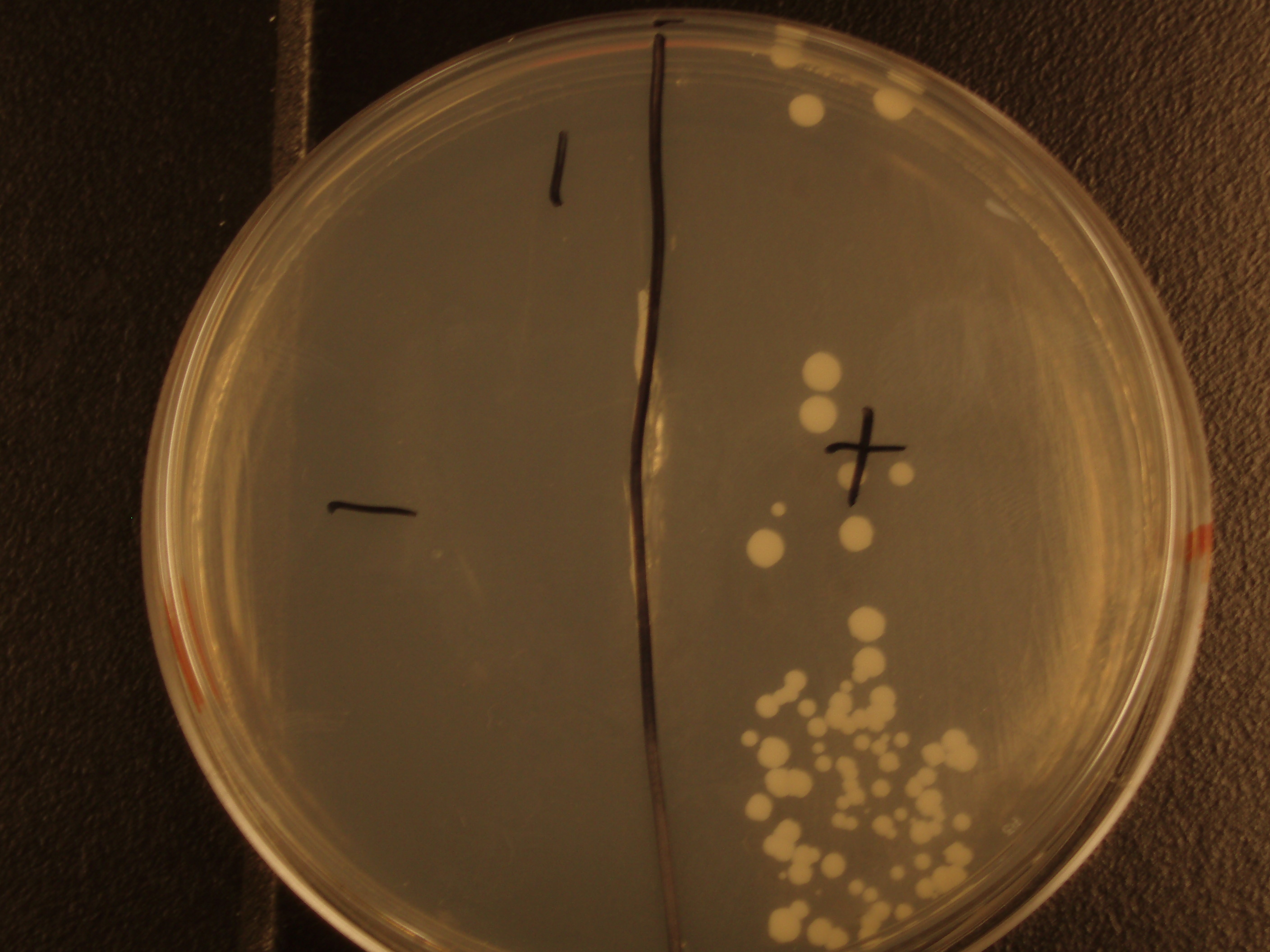
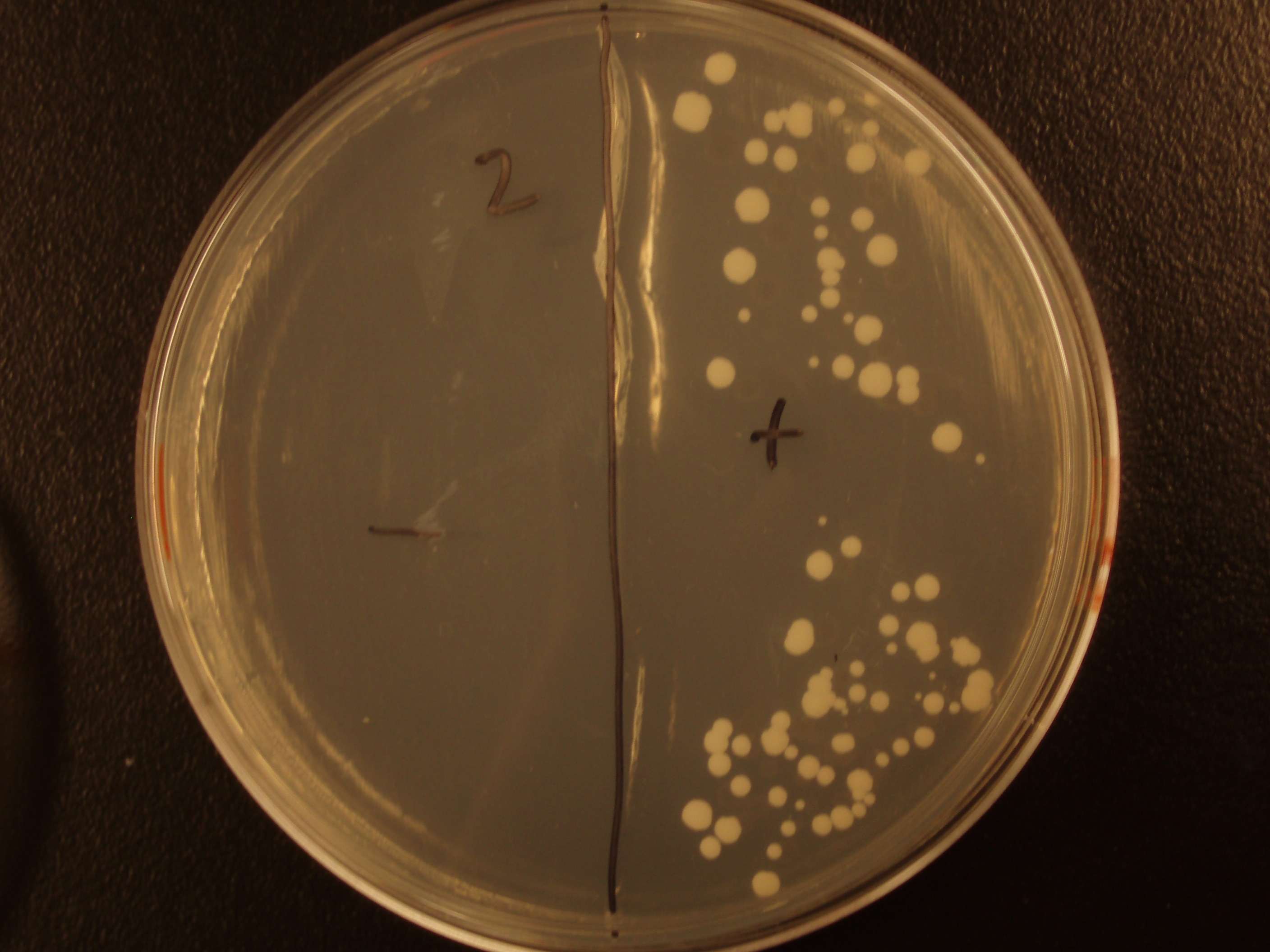
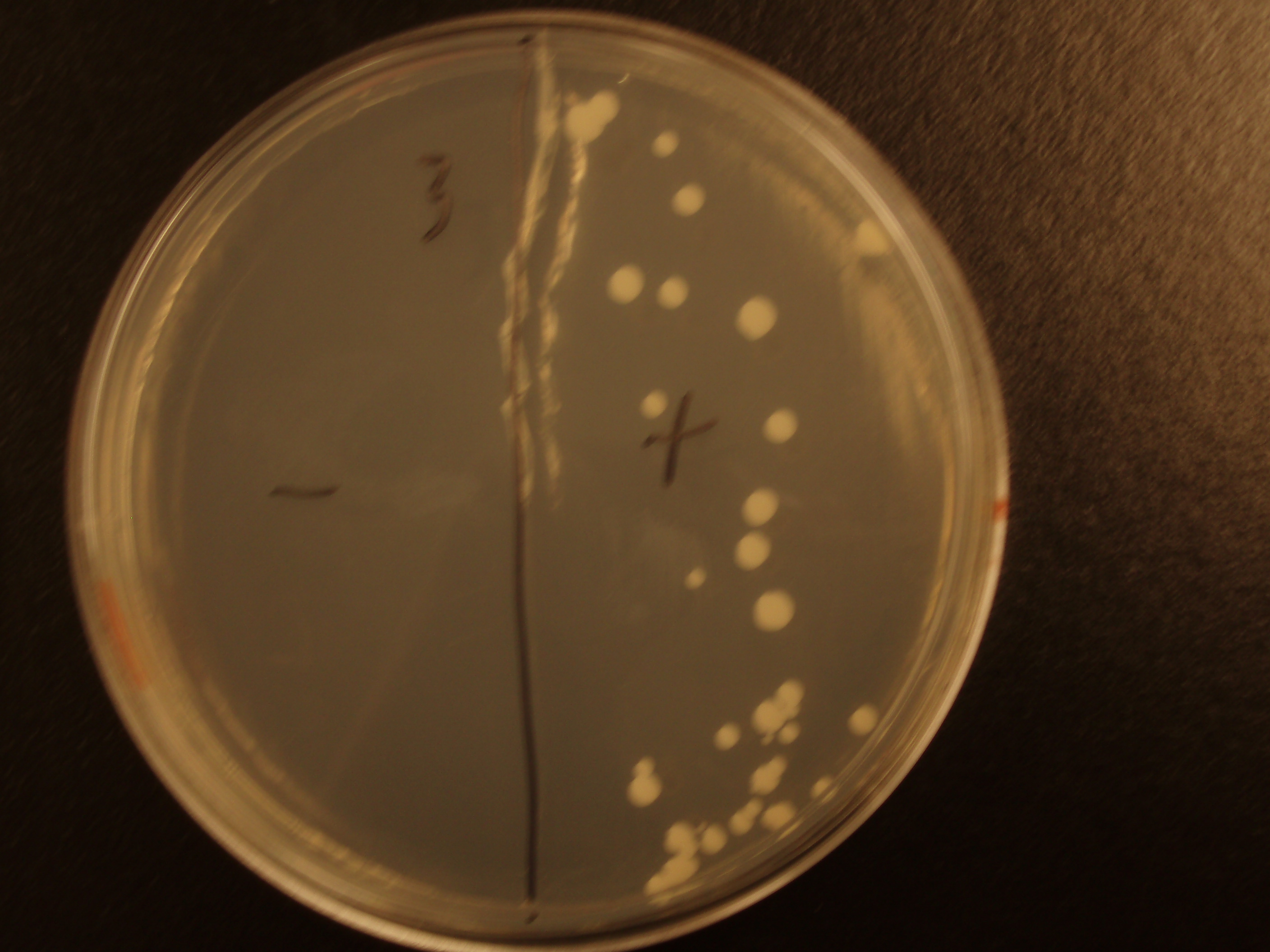
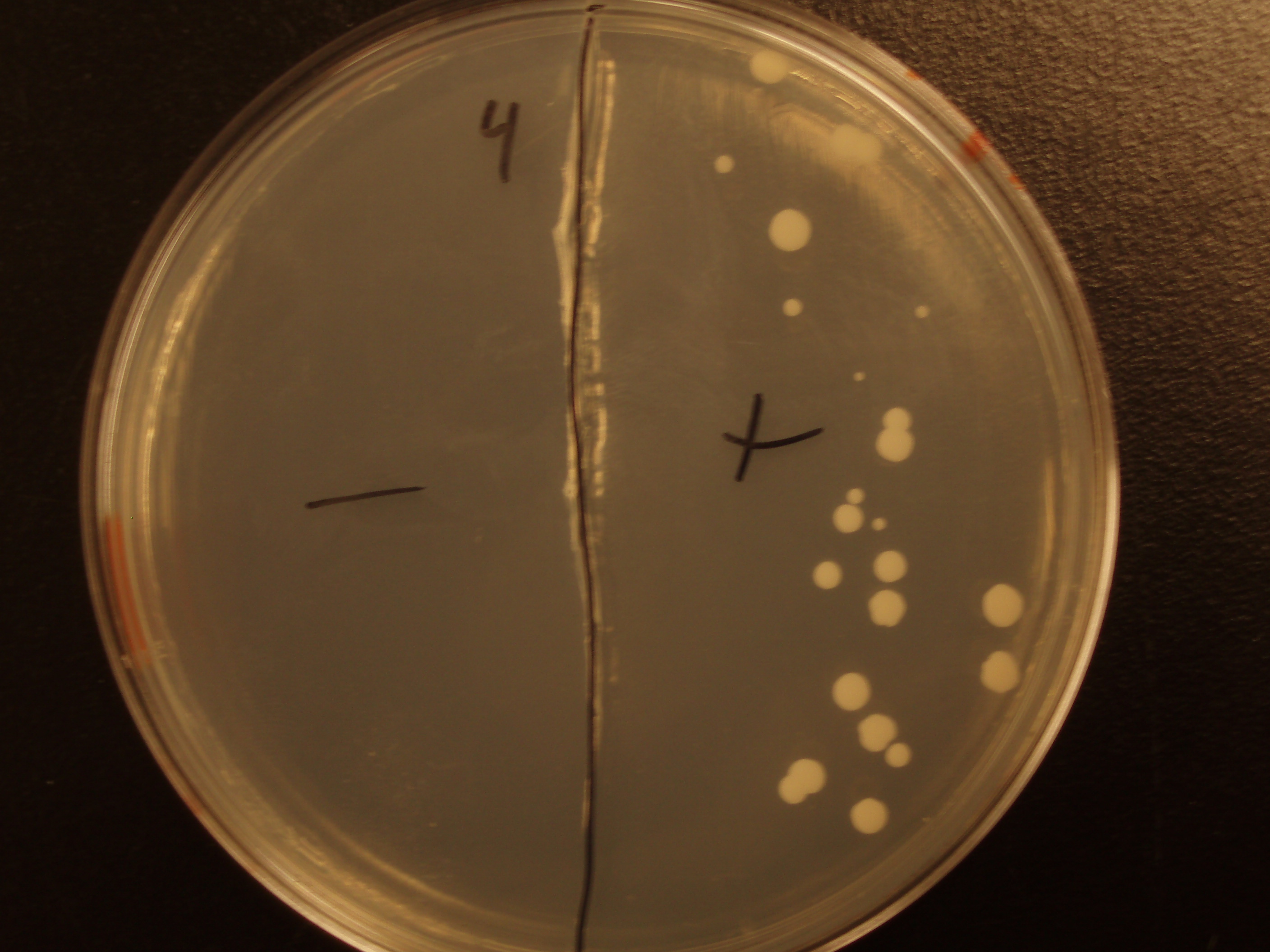
Demonstration of E. coli - S. cerevisiae Genetic Transfer
Before we could control E. coli - S. cerevisiae genetic transfer, we had to first develop a protocol and demonstrate (uncontrolled) conjugative transfer between these two organisms, shown below.
Shown are four separate experiments. In each experiment, bacteria was mixed with yeast, incubated to allow for conjugative transfer, and plated on yeast media that contains antibacterial antibiotics and lacks the essential amino acid leucine. Thus, any growth on these plates is yeast that can generate leucine. The bacteria contained a yeast shuttle plasmid with an OriT that conferred upon yeast the ability to generate leucine (right half of plates). We know that this genetic transfer occurred by conjugation since bacteria lacking the conjugative plasmid were unable to transfer the shuttle plasmid (left half of plates).
| Experiment # | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Bacteria (OD600@0.15 ≈ 3E7 bac/mL) | 250uL | 200uL | 100uL | 50uL |
| Yeast (OD600@0.1 ≈ 1E7 yeast/mL) | 250uL | 300uL | 400uL | 450uL |
| Yeast Colonies | 66 | 64 | 32 | 18 |
| Transconjugation Efficiency (Conjugation Events/Yeast Cell) | 2.64E-5 | 2.13E-5 | 8.00e-6 | 4.00e-6 |
| 
| 
| 
|
|---|
Module #4: Conferred Ability Module- Yeast Shuttle Vector
Another important part in our controlled trans-kingdom conjugation project is the yeast shuttle vector, which confers a novel ability to yeast. This plasmid is stored and maintained in E. coli, transferred to yeast via conjugation, then stored and maintained in yeast. Therefore, this yeast shuttle plasmid requires a bacterial origin of replication, a stable yeast origin of replication, a conjugative origin of transfer for RP4 mobilization, a bacterial selectable marker that confers ampicilin resistance, and a gene which imparts the ability to synthesize leucine. A plasmid containing each of these features, pAC88, was obtained from the University of Leicester where it was designed for prior research [5].
The pAC88 yeast shuttle vector, while suitable for our experiment, does not adhere to the standard BioBricks format. It contains many extraneous restriction sites strewn around the plasmid and within part sequences, while also lacking BioBrick restriction sites which flank the part sequences. Therefore, pAC88 cannot be used in standard assembly or to extract parts. In an effort to provide the synthetic biology community with a more useful, standardized yeast vector, the necessary feature for maintainance, selection, and transfer are being incorporated into the BioBrick construction base vector.
Insertion of an adaptive gene into pAC88 was also attempted. Kluyveromyces lactis, a species of milk-yeast, posesses a region of chromosomal DNA important in the production lactose digestion machinery. LAC4 and LAC12, two genes that encode for yeast galactosidase and lactose permease, respectively, are located near each other on the chromosome, and are divergently transcribed. This LAC region was amplified and inserted into an XhoI restriction site on pAC88. However, yeast transformants remained unable to digest lactose.
[1] Sprague, George F., and Jack A. Heinemann. “Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast.” Nature. Institute of Molecular Biology. University of Oregon. 340. 20 Jul 1989.
[2] Chambers P, Issaka A, Palecek SP. "Saccharomyces cerevisiae JEN1 promoter activity is inversely related to concentration of repressing sugar." Appl Environ Microbiol. 2004 Jan;70(1):8-17.
[3] Robert Sidney Cox, III, Michael G Surette, and Michael B Elowitz. "Programming gene expression with combinatorial promoters." Nature. Institute of Molecular Biology. 145. 13 November 2007
[4] Zatyka M, Jagura-Burdzy G, Thomas CM. "Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids." J Bacteriol. 1997 Dec;179(23):7201-9.
[5] Cashmore, Annette M., Steven Bates, and Brian M. Wilkins. “IncP Plasmids Are Unusually Effective in Mediating Conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 Mating System.” Journal of Bacteriology. Dec 1998. pp 6538-6543.
 "
"