Team:Wisconsin/Project
From 2008.igem.org
(→References) |
(→References) |
||
| (132 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html><link rel="stylesheet" href="https://mywebspace.wisc.edu/jluo1/wisconsinstyle.css" type="text/css"></html> | ||
| + | |||
<div style="background: #000; padding: 10px;"> | <div style="background: #000; padding: 10px;"> | ||
<div style="width:800px; height:147px; border:0px; margin:0px;">[[Image:Igemwibanner.gif]]</div> | <div style="width:800px; height:147px; border:0px; margin:0px;">[[Image:Igemwibanner.gif]]</div> | ||
| - | |||
| - | |||
__NOTOC__ | __NOTOC__ | ||
<div class="NavBar"> | <div class="NavBar"> | ||
| Line 20: | Line 20: | ||
==Abstract== | ==Abstract== | ||
<br><br> | <br><br> | ||
| - | The global fuel crisis impacts our economy, national security, and environment. The need for alternative fuels is of utmost importance. Team Wisconsin used ''Escherichia coli'' to produce biofuel precursors in an effort to find these alternative fuel sources. One project focused on using ''E. coli'' to efficiently produce sorbitol, a biofuel precursor. Using computer modeling, we determined a way to funnel glycolysis’ intermediates towards the production of sorbitol via sorbitol-6-phosphate dehydrogenase. Wisconsin’s second aim was to isolate high-energy precursors from the plant cell wall through ''E. coli'' mediated breakdown of cell wall lignin. This was achieved by inserting the gene encoding lignin peroxidase , found in the white rot fungus Phanerochaete chrysosporium, into genetically modified ''E. coli'', capable of producing and exporting the enzyme. Both projects improve current methods in the production of alternative fuels via two different, unique routes, and have the potential to move sustainable biofuel research forward. | + | The global fuel crisis impacts our economy, national security, and environment. The need for alternative fuels is of utmost importance. Team Wisconsin used ''Escherichia coli'' to produce biofuel precursors in an effort to find these alternative fuel sources. One project focused on using ''E. coli'' to efficiently produce sorbitol, a biofuel precursor. Using computer modeling, we determined a way to funnel glycolysis’ intermediates towards the production of sorbitol via sorbitol-6-phosphate dehydrogenase. Wisconsin’s second aim was to isolate high-energy precursors from the plant cell wall through ''E. coli'' mediated breakdown of cell wall lignin. This was achieved by inserting the gene encoding lignin peroxidase , found in the white rot fungus ''Phanerochaete chrysosporium'', into genetically modified ''E. coli'', capable of producing and exporting the enzyme. Both projects improve current methods in the production of alternative fuels via two different, unique routes, and have the potential to move sustainable biofuel research forward. |
==Introduction== | ==Introduction== | ||
<br> | <br> | ||
| - | + | Currently in the United States the biofuel of choice is ethanol produced from corn. Only the starch within the kernel is fermented despite the whole plant being harvested. The ratio of energy input to energy output is 1:1.34, which is inefficient for a system that is being used as a fuel source. Cellulose fermentation seems the obvious next step in biofuel production since cellulose is about 33% of all plant matter and the most abundant organic compound on earth. This has been recently accomplished by several individuals and organizations with industrial production still in the works. Advances in cellulosic biofuel technology are predicted to create an energy content of up to four billion barrels of crude oil, which accounts for 65% of oil consumption in America. Government support is assisting this transition away from fossil fuels- in 2007, US Department of Energy announced that $385 million in grants will be provided to fund six different cellulosic ethanol plants. Further support from private institutions and constant scientific advances in biofuel production methods will provide better alternatives and a change that is desperately needed. Due to the increased costs of fossil fuels, it has now become imperative to harness renewed energy through biofuel synthesis. | |
<br><br> | <br><br> | ||
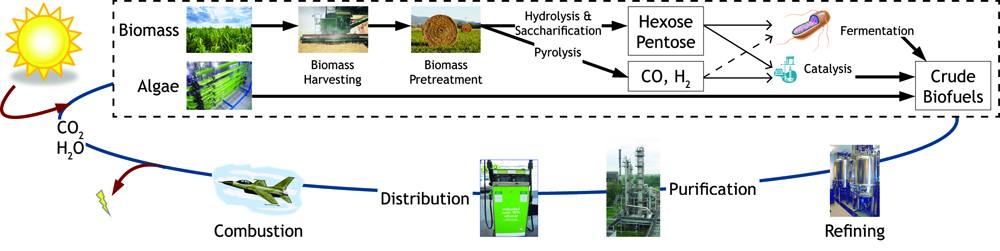
| - | [[Image:Sustain.jpg| | + | [[Image:Sustain.jpg|950px|center]]<br> |
| - | <i><b>Figure 1:</b> Map of forms of sustainable chemical production.< | + | <i><b>Figure 1:</b> Map of forms of sustainable chemical production.</i><br><br> |
| - | + | ||
| - | + | ||
==Overall Project== | ==Overall Project== | ||
| - | Fuel | + | Fuel production has come to the forefront as an important political and biological issue. It has lead to innovative pursuits of renewable fuel as well as the controversial exploitation of natural resources. Currently ethanol is the commercial biofuel of choice; however, current production and distillation of ethanol in the United States is inefficient. With this problem in mind, the iGEM Wisconsin team is looking for alternative ways to make not only ethanol, but other biofuels through synthetic biology. We've designed the following two projects based around using ''E. coli'' to produce biofuels. <br><br> |
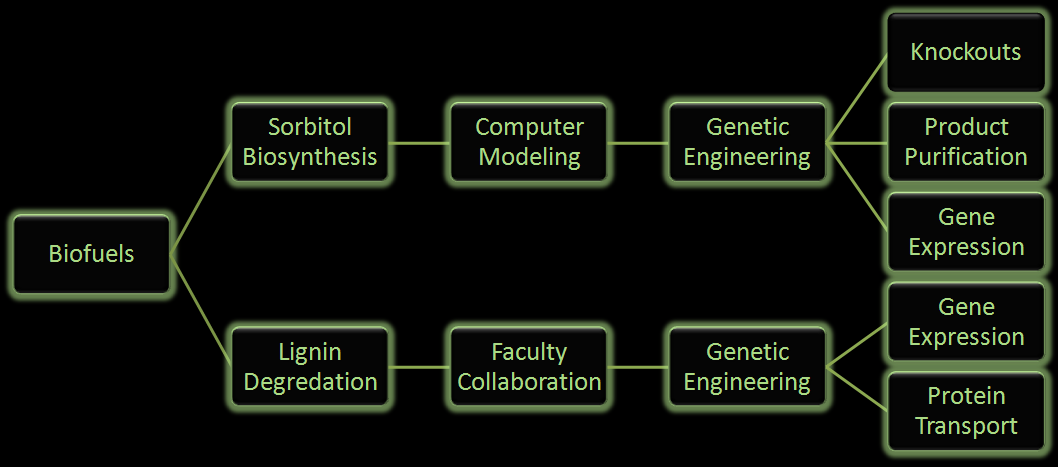
[[Image:Flowchizart.png|600px|center]] | [[Image:Flowchizart.png|600px|center]] | ||
| - | <i><b>Figure | + | <i><b>Figure 2:</b> Flow chart illustrates the process our team followed in creating our genetically engineered cells.</i> |
<br> | <br> | ||
| Line 47: | Line 45: | ||
==Lignin Peroxidase== | ==Lignin Peroxidase== | ||
<br> | <br> | ||
| - | + | Lignin is the second most abundant organic compound on earth. It is a very complex, polymorphic phenyl-propane polymer. Lignin is considered one of the hardest organic polymers to degrade due to its insolubility, high molecular weight, non-linear and non-uniform structure. Only about 10% of lignin is phenolic, meaning that ether linkages must be cleaved to break it down.<br><br> | |
| - | <br><br> | + | [[Image:Lignin_structural_image.jpg|300px]]<br> |
| - | However, there are some difficulties with the implementation. Lignin is an extremely complex molecule formed by radical reactions, and it has proved very difficult to study. The mechanism of lignin peroxidase is not fully understood. Also, there could be other genes involved with lignin breakdown some of which may not even be known to the experts. | + | ''<b>Figure 3:</b> A proposed structure of lignin.'' (Taken from: [http://www.research.uky.edu/odyssey/winter07/green_energy.html The University of Kentucky])<br><br> |
| + | Lignin, cellulose and hemi-cellulose are the three main components of the cell well. Lignin fills the space between cellulose and hemi-cellulose covalently bonding repeatedly to both. This hinders cell wall breakdown and in nature prevents access to cellulose as a substrate for enzymes. <br><br> | ||
| + | [[Image:Complete_cell_wall_image.jpg|215px]][[Image:Cellulose_(cell_wall_complex)_image.jpg|215px]][[Image:Hemicellulose_(cell_wall_complex)_image.jpg|215px]][[Image:Lignin_(cell_wall_complex)_image.jpg|215px]]<br> | ||
| + | ''<b>Figure 4:</b> These images show the different components of the plant cell wall complex: cellulose(orange), hemi-cellulose(blue), and lignin(green).'' (Taken from: [http://www.ceres.net/AboutUs/AboutUs-Biofuels-Carbo.html Ceres: the energy crop company])<br><br> | ||
| + | There are enzymes and organisms out there that can break down lignin. Adding genes necessary for the enzymatic breakdown of lignin to an easily grown and manipulated microbe, such as ''E. coli'', allows for the breakdown of lignin on a large scale for the purpose of biofuel production. Lignin degradation would result in greater cellulosic fermentation efficiency by allowing easier access to cellulose and hemi-cellulose. The by products of lignin degradation could also be converted into a fuel, tapping yet another untapped abundant energy source. The white rot fungus, ''Phanerochaete chrysosporium'', has the enzyme lignin peroxidase which seems to be promising for our purpose. If successful, a wide range of more efficient biofuel production methods will become available. This would not only result in increased fermentation efficiency of ethanol, but of various other fuels as well. Corn would also no longer have to be used as the plant source, eliminating much of the controversy and inefficiency from the process. Switch grass, wood scraps, yard waste, or just about any other plant matter could potentially be turned into fuel.<br><br> | ||
| + | However, there are some difficulties with the implementation. Lignin is an extremely complex molecule formed by radical reactions, and it has proved very difficult to study. The mechanism of lignin peroxidase is not fully understood. Also, there could be other genes involved with lignin breakdown, some of which may not even be known to the experts. | ||
<br><br> | <br><br> | ||
[[Image:Final_test_sphere_try_act.png|Lignin Peroxidase|400px]] | [[Image:Final_test_sphere_try_act.png|Lignin Peroxidase|400px]] | ||
[[Image:Final_test_whole_lip.png|Lignin Peroxidase|400px]] | [[Image:Final_test_whole_lip.png|Lignin Peroxidase|400px]] | ||
<br> | <br> | ||
| - | <i><b>Figure | + | <i><b>Figure 5:</b> Images of Lignin peroxidase. Disulfide bonds (yellow), Ca2+ ions (green spheres), Heme access |
channel (purple), Surface active cite (red) with hydroxylated typ171 | channel (purple), Surface active cite (red) with hydroxylated typ171 | ||
(orange), Hydrogen peroxide active cite (tan wireframe, above heme), | (orange), Hydrogen peroxide active cite (tan wireframe, above heme), | ||
| - | Heme group (CPK coloring space fill.)</i> | + | Heme group (CPK coloring space fill.) These images and video were made with the PDB file 1QPA on pymol.<br>[http://www.youtube.com/watch?v=dYdru4ig5Q4 <font color=#aada84><b> 3D Video of Lignin peroxidase.</b></font>]</i> |
| - | <br> | + | <br><br> |
| + | |||
| + | <html> | ||
| + | <div> | ||
| + | <!--YouTube--> | ||
| + | <div align="center" class="youtube"> | ||
| + | <object width="425" height="344"><param name="movie" | ||
| + | value="http://www.youtube.com/v/dYdru4ig5Q4&hl=en&fs=1&rel=0"></param><param | ||
| + | name="allowFullScreen" value="true"></param><embed | ||
| + | src="http://www.youtube.com/v/dYdru4ig5Q4&hl=en&fs=1&rel=0" | ||
| + | type="application/x-shockwave-flash" allowfullscreen="true" | ||
| + | width="425" height="344"></embed></object> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | The above images and video give the general features of lignin peroxidase (''lip''), which includes 4 disulfide bonds and 2 Ca2+ that are conserved through all ''lip''. The heme access channels are divided into a larger hydrophobic channel and a smaller hydrophilic channel. Hydrogen peroxide must travel through the hydrophilic channel since the heme is embedded inside of the protein. At the end of the channel, hydrogen peroxide turns into water by oxidizing 2 electrons from the heme at the hydrogen peroxide active site. This allows ''Lip'' to oxidize substrates (veratryl alcohol for example.) Since bulky molecules are not able to reduce the embedded heme directly, they must be oxidized at the surface. A Tryptophan with a hydroxyl group on its beta carbon is the center of the surface active site and allows the substrate's electrons to transfer through the protein into the center and reduce the heme, allowing the whole system to start over again. However, much is still unknown about ''Lip'', such as the exact mechanism for lignin degradation or veratryl alcohol's precise role in increasing efficiency. | ||
| + | |||
| + | In researching for an appropriate assay in analyzing the amount of lignin peroxidase produced by our ''E. coli'' strain, we found that the veratryl alcohol assay, developed in 1984, is currently the most widely used. It works by viewing the absorbance spectrum of the reaction solution. As veratryl alcohol is oxidized by lignin peroxidase over time, the optical density (OD) and wavelength of the absorbance peak change. By observing the disappearance of the substrate (the rate of change), we can determine the amount of active enzyme in the solution. | ||
| + | <br><br> | ||
| + | In doing additional research we came across a newer and seemingly more effective assay—the Azure B assay. Research showed that Azure B (a blue textile dye) has a higher specificity to lignin peroxidase compared to veratryl alcohol, as veratryl alcohol can be oxidized by fungal enyzmes other than lignin peroxidase. The Azure B assay works the same way as the veratryl alcohol assay in that the disappearance of the dye is measured to determine the amount of lignin peroxidase. The reaction solution comprises of a tartrate buffer (50 mM at pH 4.5), the dye Azure B (32 μM), hydrogen peroxide (100 μM), and the enzyme to be measured, lignin peroxidase. | ||
| + | <br><br> | ||
| + | |||
| + | [[Image:Azureb.jpg|300px|center]] | ||
| + | <i><b>Figure 6:</b></i> [http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/A4043 <font color=#aada84>Azure B. Sigma Aldrich.</font>] <i>(2008). Retrieved Oct 18.</i> | ||
| + | |||
| + | <br><br> | ||
| + | Lignin peroxidase has a few disulfide bonds, which are unable to form within ''E. coli''. To combat this problem we used a strain (any of the Origami strains) available from Novagen that is a knockout for both thioredoxin reductase and glutathione reductase. This prevents the cell from reducing disulfide bonds as they form, allowing for eukaryotic proteins containing disulfide bonds to be expressed in the active form. | ||
| + | <br><br> | ||
| + | Since the extremely large plant cell wall complex cannot be imported into the cell we had to figure out a way to get lignin peroxidase out of the cell. This could either be done by lysing the cell and recovering the enzyme or by exporting the enzyme from the cell using a native export system. We looked into using two native export pathways in ''E. coli'', the Sec pathway and the Twin Arginine Translocation (Tat) pathway. Proteins are recognized for transportation across the membrane by signal peptide sequences. The Sec pathway exports unfolded proteins, which then fold once in the periplasm. The Tat pathway exports already folded proteins. Since the cytoplasm within the Origami strains was an appropriate place for the protein to fold correctly we focused on the Tat pathway. Export of lignin peroxidase could be achieved by fusing sequences encoding for Tat signal peptides in frame to the lignin peroxidase gene.<br><br> | ||
| + | |||
| + | [[Image:Export.jpg|500px|center]] | ||
| + | <i><b>Figure 7:</b>TAT pathways can export entire folded proteins, while Sec involves an unfolding step, export of the unfolded protein and a refolding of the unfolded protein. Both pathways result in a folded protein in the periplasm. </i> | ||
| + | |||
| + | |||
| + | Another difficulty is the crucial addition of the heme group. Producing lignin peroxidase will only give us nonfunctional polypeptides if we do not have enough heme. We decided to focus up regulation on aminolevulinic acid (ALA) synthesis since it is the rate limiting step for heme production. However, instead of the time consuming up regulation of enzymes responsible for ALA, we decided to simply add ALA to the media, which would then be taken up by e coli. This chemical approach cuts out the rate limiting step entirely and is a cheap method that is realistic for the short time scale of iGEM research. | ||
==Sorbitol Biosynthesis== | ==Sorbitol Biosynthesis== | ||
| Line 65: | Line 103: | ||
Sorbitol anabolism is part of the glycolysis pathway in ''E.coli''. In the pathway, interconversion between fructose and sorbitol is catalyzed by sorbitol dehydrogenase(SDH), a monomeric enzyme that uses NADH in the process. Natural expression of SDH is generally up-regulated only in the presence of sorbitol as a carbon source. The initial idea to stimulate sorbitol over-production was to simply insert a plasmid containing the gene which codes for SDH and grow the cells in an environment lacking sorbitol thus pushing the metabolic reaction governing fructose and sorbitol interconversion towards sorbitol. | Sorbitol anabolism is part of the glycolysis pathway in ''E.coli''. In the pathway, interconversion between fructose and sorbitol is catalyzed by sorbitol dehydrogenase(SDH), a monomeric enzyme that uses NADH in the process. Natural expression of SDH is generally up-regulated only in the presence of sorbitol as a carbon source. The initial idea to stimulate sorbitol over-production was to simply insert a plasmid containing the gene which codes for SDH and grow the cells in an environment lacking sorbitol thus pushing the metabolic reaction governing fructose and sorbitol interconversion towards sorbitol. | ||
| - | The gene encoding for SDH, ''srlD'', is part of the ''srl'' | + | <br><br> |
| + | [[Image:Sorbitol Dehydrogenase fireframe ray traced.png|500px|center]] | ||
| + | <br> | ||
| + | <i><b>Sorbitol Dehydrogenase</b> NADH binding site (purple wire frame), sorbitol active site (red wire frame), sorbitol hydrogen bonding amino acids (orange wire frame, doesn't include hydrogen bonding from the sorbitol active site) This image was made with PDB file 1K2W on pymol.</i> | ||
| + | |||
| + | |||
| + | The gene encoding for SDH, ''srlD'', is part of the ''srl'' operon in ''E.coli'' (operon pictured below). Information on the SDH assembly pathway and enzymatic activity is limited. Consequently we decided that insertion of both the ''srlD'' gene and ''srl'' operon into vectors would take more time initially but would potentially be beneficial due to the chance that the gene alone would not result in effective SDH production. | ||
| - | [[Image:SRLOp.JPG| | + | [[Image:SRLOp.JPG|950px|SRL Operon|center]] |
| - | <br><i><b>Figure | + | <br><i><b>Figure 6:</b> SRL Operon</i><br><br> |
Increased cellular production of SDH would most likely increase the amount of sorbitol produced by the cell, however it could not alone account for the type of mass production that would make sorbitol biosynthesis commercially useful. In order to determine other methods of increasing sorbitol production, our group turned to computer modeling. | Increased cellular production of SDH would most likely increase the amount of sorbitol produced by the cell, however it could not alone account for the type of mass production that would make sorbitol biosynthesis commercially useful. In order to determine other methods of increasing sorbitol production, our group turned to computer modeling. | ||
| - | |||
<br> | <br> | ||
| + | <span style="font-size:16px">'''Phosphofructokinase Knockout'''</span> | ||
| - | + | [[Image:PFK-Knockout-wiki.png|350px|PFK Knockout|center]] | |
| + | <br><i><b>Figure 7:</b> PFK Knockout in the metabolic pathway.</i><br><br> | ||
| + | The idea behind a phosphofructokinase (PFK) knockout is to literally stop the glycolysis pathway after fructose-6-phosphate. The backup of metabolic products caused by the knockout should pool and form a favorable pathway for increased sorbitol production. The obvious problem with this procedure is that knocking out a part of the glycolysis pathway under normal circumstances kills the cell. This is also why modeling didn't prove useful as the program showed the cell would be dead. We set out to determine how to bypass this normal cellular function in order to use it to increase sorbitol production. | ||
| + | Experiments have been successfully done to create a viable PFK chromosomal knockouts that contain inducible plasmids encoded with the ''pfk'' gene. After obtaining this strain we have done countless growth curves with varying media to create ideal conditions for cell growth. After the optimal media is obtained, the addition of a plasmid containing ''srlD'' or the ''srl'' operon will be introduced to create what we hope will be this metabolic pathway(right). | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <span style="font-size:16px">'''Triose Phosphate Isomerase Knockout'''</span> | ||
| - | [[Image:TPI-Knockout.png| | + | [[Image:TPI-Knockout.png|350px|TPI Knockout|center]] |
| - | Computer modeling was done to determine gene knockouts that increased cellular flux towards sorbitol production. The model was given to us by Dr. | + | <br><i><b>Figure 8:</b> TPI Knockout in the metabolic pathway.</i><br><br> |
| + | Computer modeling was done to determine gene knockouts that increased cellular flux towards sorbitol production. The model was given to us by Dr. Jennie Reed of University of Wisconsin-Madison. More extensive information about modeling can be found at our [[Team:Wisconsin/Modeling|<font color=#aada84><b>modeling</b></font>]] page. Through the use of our model we determined that knocking out the gene encoding for triose phosphate isomerase would be the most effective at increasing sorbitol production when combined with ''srl'' gene up-regulation. | ||
Triose phosphate isomerase(TPI) is another enzyme in ''E.coli'''s glycolysis pathway. Its function is to catalyze the interconversion of 3-Phosphoglyceraldehyde to Dihidroxy-acetone-phosphate and back. Knocking out the gene which encodes TPI stops this interconversion, a process which removes products of glycolysis from the cellular reaction mixture shifting the overall reaction downwards towards pyruvate. In essence, knocking out the TPI gene backs up glycolysis and produces a shift towards sorbitol (Pictured right). | Triose phosphate isomerase(TPI) is another enzyme in ''E.coli'''s glycolysis pathway. Its function is to catalyze the interconversion of 3-Phosphoglyceraldehyde to Dihidroxy-acetone-phosphate and back. Knocking out the gene which encodes TPI stops this interconversion, a process which removes products of glycolysis from the cellular reaction mixture shifting the overall reaction downwards towards pyruvate. In essence, knocking out the TPI gene backs up glycolysis and produces a shift towards sorbitol (Pictured right). | ||
| Line 88: | Line 141: | ||
Even with the promising modeling results, our group worked on alternative routes for increased cellular sorbitol. One of these routes, knocking out phosphofructokinase, was an additional project that our group undertook. | Even with the promising modeling results, our group worked on alternative routes for increased cellular sorbitol. One of these routes, knocking out phosphofructokinase, was an additional project that our group undertook. | ||
| - | <br> | + | <span style="font-size:16px">'''Approach'''</span><br><br> |
| - | <br> | + | <br>[[Image:Srldgene.jpg|600px|SrlD|center]] |
| + | <br><i><b>Figure 9:</b> 1. Wild Type E.coli MG1655 is grown on an LB plate<br> | ||
| - | < | + | 2. A colony is selected and grown overnight<br> |
| - | + | 3. The chromosomal DNA is harvested from the over night culture<br> | |
| - | + | 4. srlD gene is cloned and amplified via a PCR reaction adding restriction sites for Xba1 and Spe1 at either end<br> | |
| - | + | 5. The biobrick vector pSB1AK3 as well as the srlD PCR product are digested with Xba1 and Spe1<br> | |
| - | + | 6. The two digestion products are then ligated together<br> | |
| - | + | 7. The ligation product is then transformed into competent E.coli<br> | |
| + | 8. E. coli is then plated on LB plates containing Amp/Kan to select for transformants</i><br><br> | ||
==References== | ==References== | ||
| Line 103: | Line 158: | ||
<br>Lorenzo Nissen, Gaspar Pérez-Martínez, María J. Yebra (2005) Sorbitol synthesis by an engineered Lactobacillus casei strain expressing a sorbitol-6-phosphate dehydrogenase gene within the lactose operon �FEMS Microbiology Letters 249 (1) , 177–183 doi:10.1016/j.femsle.2005.06.010 | <br>Lorenzo Nissen, Gaspar Pérez-Martínez, María J. Yebra (2005) Sorbitol synthesis by an engineered Lactobacillus casei strain expressing a sorbitol-6-phosphate dehydrogenase gene within the lactose operon �FEMS Microbiology Letters 249 (1) , 177–183 doi:10.1016/j.femsle.2005.06.010 | ||
<br>Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. | <br>Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. | ||
| - | <br>Ladero, V., Ramos, A., Wiersma, A., Goffin, P., Schanck, A., Kleerebezem, M., | + | <br>Ladero, V., Ramos, A., Wiersma, A., Goffin, P., Schanck, A., Kleerebezem, M., et al. (2007) High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering. Appl Environ Microbiol 73: 1864–1872. |
<br>Metzger J (2006) Production of liquid hydrocarbons from biomass. Angew. Chem. Int. Ed. 2006, 45, 696-698. | <br>Metzger J (2006) Production of liquid hydrocarbons from biomass. Angew. Chem. Int. Ed. 2006, 45, 696-698. | ||
<br>Lovingshimera, M. R., Siegeleb, D., & Reinharta, G. D. (2006). Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources. Protein Expression and Purification , 46 (2), 475-482. | <br>Lovingshimera, M. R., Siegeleb, D., & Reinharta, G. D. (2006). Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources. Protein Expression and Purification , 46 (2), 475-482. | ||
<br>Karacaoğlan, V., & Özer, I. (2005). Steady-state kinetic properties of sorbitol dehydrogenase from chicken liver. Comparative Biochemistry and Physiology , 140, 309-312. | <br>Karacaoğlan, V., & Özer, I. (2005). Steady-state kinetic properties of sorbitol dehydrogenase from chicken liver. Comparative Biochemistry and Physiology , 140, 309-312. | ||
| - | <br>Archibald, F. S. A new assay for lignin-type peroxidases employing the dye azure B. Applied and Environmental Microbiology, 58(9), 3110-3116. | + | <br>Archibald, F. S. "A new assay for lignin-type peroxidases employing the dye azure B." Applied and Environmental Microbiology, 58(9), 3110-3116. |
| - | + | <br>"New Method Rapidly Produces Low-Cost Biofuels From Wood, Grass". Energy Daily (2008-04-14).<br> | |
| - | </ | + | The International Energy Agency. 26 Aug. 2008. Biofuels for Transport: An International Perspective. April 2004. The International Energy Agency. 26 Aug. 2008. <http://www.iea.org/textbase/nppdf/free/2004/biofuels2004.pdf><br> |
| + | Doyle, Wendy A., Andrew T. Smith. “Expression of lignin peroxidase H8 in Escherichia coli: folding and activation of the recombinant enzyme with Ca2+ and haem.” Biochemistry Journal 315 (1996): 15-19.<br> | ||
| + | Hammel, Kenneth E., Dan Cullen. "Role of fungal peroxidases in biological ligninolysis." Current Opinion in Plant Biology 11 (2008): 349-355.<br> | ||
| + | "Cellulose." Encyclopedia Britannica. 2008. Encyclopedia Britannica Online. 28 Oct. 2008.<br> | ||
| + | DeLisa, Matthew P., Danielle Tullman, and George Georgiou. "Folding quality in the export of proteins by the bacterial twin arginine translocation pathway." Proceedings of the National Academy of Sciences 100 (2003): 6155-6120.<br> | ||
| + | Tullman-Ereck, Danielle, Matthew P. DeLisa, Yasuaki Kawarasaki, Pooya Iranpour, Brian Ribnicky, Tracy Palmer, and George Georgiou. "Export Pathway Selectivity of Escherichia Coli Twin Arginine Translocation Signal Peptides." Journal of Biological Chemistry 282 (2007): 8309-83116.<br> | ||
| + | Brenda: the Comprehensive Enzyme Information System. 2008.2. Technical University of Braunschweig Department of Bioinformatics and Biochemistry. 2 July 2008. <http://www.brenda-enzymes.info/><br> | ||
| + | hello<br> | ||
| + | Guzman L., Belin D., CarsonM., and Beckwith L. (1995). Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. Journal of Bacteriology. 177(14): 4121-4130. | ||
| + | Thomas Choinowski, Wolfgang Blodig, Kaspar H. Winterhalter and Klaus piontek (1999) The crystal structure of lignin peroxidase at 1.70A resolution reveals a hydrozy group on the Cb of tryptophan171: A novel radical site formed during the redox cycle. Journal of Molecular Biology 286, 809-827 | ||
| + | Thomas L Poulos, Steven L Edwards, Hiroyuki Wariishii, and Michael H Gold (1993) Crystallographic reginement of lignin peroxidase at 2A. The journal of biological chemistry 268, 4429-4440 | ||
| + | Wendy A Doyel, Wolfgang Blodig, Nigel C Veitch, Klaus Piontek and Andrew T smith (1998) Two substrate intereaction sites in lignin peroxidas e revealed by site directed mutagenesis. Biochemistry 37, 15097-15105 | ||
| + | Wolfgang Blodig, Andrew T smith, Wendy A dDoyel, and Klaus Piontek (2001) Crystal Structures of Pristiena dn oxidatively processed lignin peroxidase expressed in escherichia coli and of the w171f variant that eliminates the redox activite tryptophan 171. implications for the reaction mechanisim. Journal Molecular Biology (2001) 305, 851-861 | ||
Latest revision as of 03:58, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Abstract
The global fuel crisis impacts our economy, national security, and environment. The need for alternative fuels is of utmost importance. Team Wisconsin used Escherichia coli to produce biofuel precursors in an effort to find these alternative fuel sources. One project focused on using E. coli to efficiently produce sorbitol, a biofuel precursor. Using computer modeling, we determined a way to funnel glycolysis’ intermediates towards the production of sorbitol via sorbitol-6-phosphate dehydrogenase. Wisconsin’s second aim was to isolate high-energy precursors from the plant cell wall through E. coli mediated breakdown of cell wall lignin. This was achieved by inserting the gene encoding lignin peroxidase , found in the white rot fungus Phanerochaete chrysosporium, into genetically modified E. coli, capable of producing and exporting the enzyme. Both projects improve current methods in the production of alternative fuels via two different, unique routes, and have the potential to move sustainable biofuel research forward.
Introduction
Currently in the United States the biofuel of choice is ethanol produced from corn. Only the starch within the kernel is fermented despite the whole plant being harvested. The ratio of energy input to energy output is 1:1.34, which is inefficient for a system that is being used as a fuel source. Cellulose fermentation seems the obvious next step in biofuel production since cellulose is about 33% of all plant matter and the most abundant organic compound on earth. This has been recently accomplished by several individuals and organizations with industrial production still in the works. Advances in cellulosic biofuel technology are predicted to create an energy content of up to four billion barrels of crude oil, which accounts for 65% of oil consumption in America. Government support is assisting this transition away from fossil fuels- in 2007, US Department of Energy announced that $385 million in grants will be provided to fund six different cellulosic ethanol plants. Further support from private institutions and constant scientific advances in biofuel production methods will provide better alternatives and a change that is desperately needed. Due to the increased costs of fossil fuels, it has now become imperative to harness renewed energy through biofuel synthesis.
Figure 1: Map of forms of sustainable chemical production.
Overall Project
Fuel production has come to the forefront as an important political and biological issue. It has lead to innovative pursuits of renewable fuel as well as the controversial exploitation of natural resources. Currently ethanol is the commercial biofuel of choice; however, current production and distillation of ethanol in the United States is inefficient. With this problem in mind, the iGEM Wisconsin team is looking for alternative ways to make not only ethanol, but other biofuels through synthetic biology. We've designed the following two projects based around using E. coli to produce biofuels.
Figure 2: Flow chart illustrates the process our team followed in creating our genetically engineered cells.
One project focuses on using E. coli to produce sorbitol, a sugar alcohol, in large quantities for eventual commercial scale catalytic conversion to hydrocarbons. Along with producing sorbitol, we've modeled alterations in E. coli to make sorbitol production from a glycerol carbon source possible. Our aim is to modify a cell that will utilize glycerol, a byproduct from biodiesel production, and effectively create sorbitol.
In the other project we are attempting to use E. coli to break down lignin from plant matter. This would increase the efficiency of biofuel production from plant biomass. We are currently aiming to insert a fungal gene coding for lignin peroxidase into E. coli. Lignin breakdown will be made possible through the transport of lignin peroxidase out of the cell.
Lignin Peroxidase
Lignin is the second most abundant organic compound on earth. It is a very complex, polymorphic phenyl-propane polymer. Lignin is considered one of the hardest organic polymers to degrade due to its insolubility, high molecular weight, non-linear and non-uniform structure. Only about 10% of lignin is phenolic, meaning that ether linkages must be cleaved to break it down.
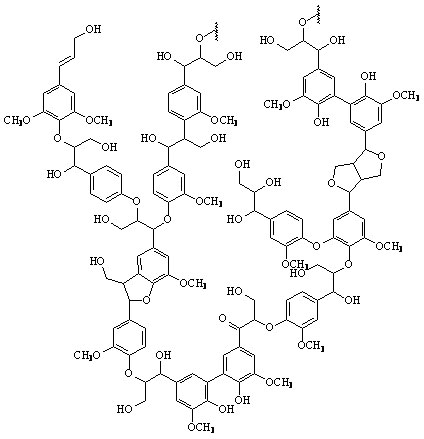
Figure 3: A proposed structure of lignin. (Taken from: [http://www.research.uky.edu/odyssey/winter07/green_energy.html The University of Kentucky])
Lignin, cellulose and hemi-cellulose are the three main components of the cell well. Lignin fills the space between cellulose and hemi-cellulose covalently bonding repeatedly to both. This hinders cell wall breakdown and in nature prevents access to cellulose as a substrate for enzymes.
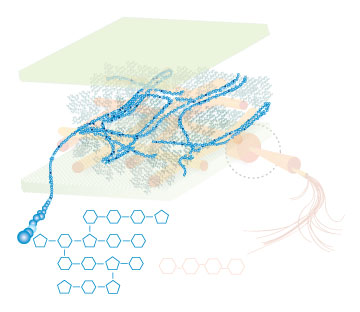
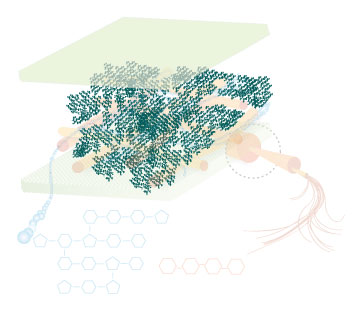
Figure 4: These images show the different components of the plant cell wall complex: cellulose(orange), hemi-cellulose(blue), and lignin(green). (Taken from: [http://www.ceres.net/AboutUs/AboutUs-Biofuels-Carbo.html Ceres: the energy crop company])
There are enzymes and organisms out there that can break down lignin. Adding genes necessary for the enzymatic breakdown of lignin to an easily grown and manipulated microbe, such as E. coli, allows for the breakdown of lignin on a large scale for the purpose of biofuel production. Lignin degradation would result in greater cellulosic fermentation efficiency by allowing easier access to cellulose and hemi-cellulose. The by products of lignin degradation could also be converted into a fuel, tapping yet another untapped abundant energy source. The white rot fungus, Phanerochaete chrysosporium, has the enzyme lignin peroxidase which seems to be promising for our purpose. If successful, a wide range of more efficient biofuel production methods will become available. This would not only result in increased fermentation efficiency of ethanol, but of various other fuels as well. Corn would also no longer have to be used as the plant source, eliminating much of the controversy and inefficiency from the process. Switch grass, wood scraps, yard waste, or just about any other plant matter could potentially be turned into fuel.
However, there are some difficulties with the implementation. Lignin is an extremely complex molecule formed by radical reactions, and it has proved very difficult to study. The mechanism of lignin peroxidase is not fully understood. Also, there could be other genes involved with lignin breakdown, some of which may not even be known to the experts.
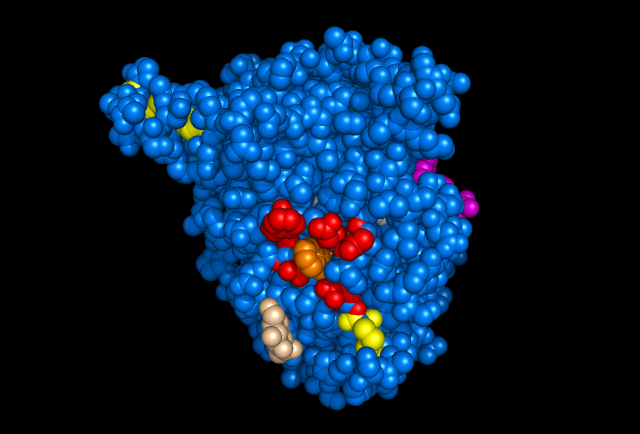
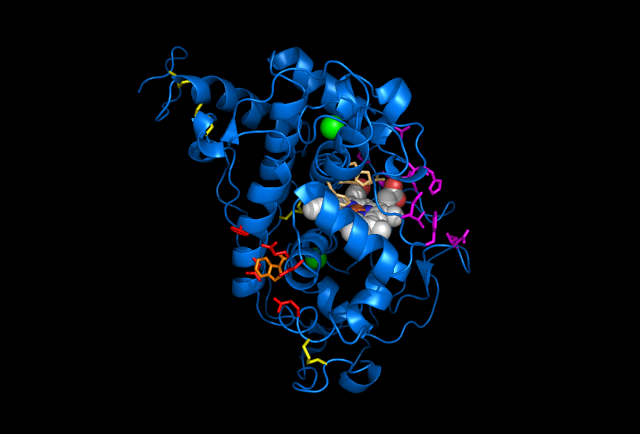
Figure 5: Images of Lignin peroxidase. Disulfide bonds (yellow), Ca2+ ions (green spheres), Heme access
channel (purple), Surface active cite (red) with hydroxylated typ171
(orange), Hydrogen peroxide active cite (tan wireframe, above heme),
Heme group (CPK coloring space fill.) These images and video were made with the PDB file 1QPA on pymol.
[http://www.youtube.com/watch?v=dYdru4ig5Q4 3D Video of Lignin peroxidase.]
The above images and video give the general features of lignin peroxidase (lip), which includes 4 disulfide bonds and 2 Ca2+ that are conserved through all lip. The heme access channels are divided into a larger hydrophobic channel and a smaller hydrophilic channel. Hydrogen peroxide must travel through the hydrophilic channel since the heme is embedded inside of the protein. At the end of the channel, hydrogen peroxide turns into water by oxidizing 2 electrons from the heme at the hydrogen peroxide active site. This allows Lip to oxidize substrates (veratryl alcohol for example.) Since bulky molecules are not able to reduce the embedded heme directly, they must be oxidized at the surface. A Tryptophan with a hydroxyl group on its beta carbon is the center of the surface active site and allows the substrate's electrons to transfer through the protein into the center and reduce the heme, allowing the whole system to start over again. However, much is still unknown about Lip, such as the exact mechanism for lignin degradation or veratryl alcohol's precise role in increasing efficiency.
In researching for an appropriate assay in analyzing the amount of lignin peroxidase produced by our E. coli strain, we found that the veratryl alcohol assay, developed in 1984, is currently the most widely used. It works by viewing the absorbance spectrum of the reaction solution. As veratryl alcohol is oxidized by lignin peroxidase over time, the optical density (OD) and wavelength of the absorbance peak change. By observing the disappearance of the substrate (the rate of change), we can determine the amount of active enzyme in the solution.
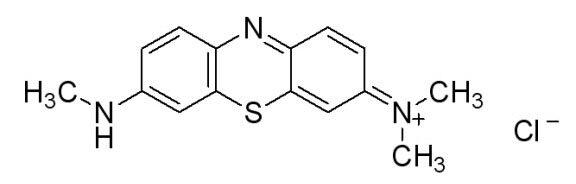
In doing additional research we came across a newer and seemingly more effective assay—the Azure B assay. Research showed that Azure B (a blue textile dye) has a higher specificity to lignin peroxidase compared to veratryl alcohol, as veratryl alcohol can be oxidized by fungal enyzmes other than lignin peroxidase. The Azure B assay works the same way as the veratryl alcohol assay in that the disappearance of the dye is measured to determine the amount of lignin peroxidase. The reaction solution comprises of a tartrate buffer (50 mM at pH 4.5), the dye Azure B (32 μM), hydrogen peroxide (100 μM), and the enzyme to be measured, lignin peroxidase.
Figure 6: [http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/A4043 Azure B. Sigma Aldrich.] (2008). Retrieved Oct 18.
Lignin peroxidase has a few disulfide bonds, which are unable to form within E. coli. To combat this problem we used a strain (any of the Origami strains) available from Novagen that is a knockout for both thioredoxin reductase and glutathione reductase. This prevents the cell from reducing disulfide bonds as they form, allowing for eukaryotic proteins containing disulfide bonds to be expressed in the active form.
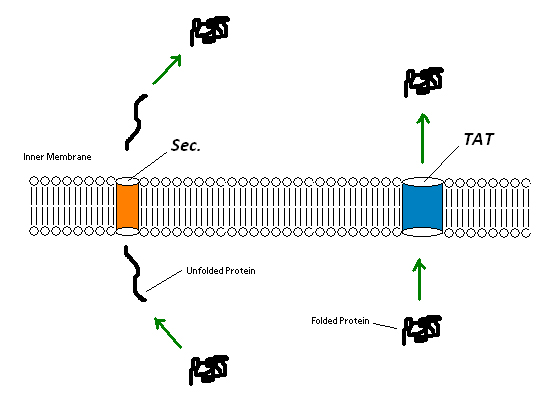
Since the extremely large plant cell wall complex cannot be imported into the cell we had to figure out a way to get lignin peroxidase out of the cell. This could either be done by lysing the cell and recovering the enzyme or by exporting the enzyme from the cell using a native export system. We looked into using two native export pathways in E. coli, the Sec pathway and the Twin Arginine Translocation (Tat) pathway. Proteins are recognized for transportation across the membrane by signal peptide sequences. The Sec pathway exports unfolded proteins, which then fold once in the periplasm. The Tat pathway exports already folded proteins. Since the cytoplasm within the Origami strains was an appropriate place for the protein to fold correctly we focused on the Tat pathway. Export of lignin peroxidase could be achieved by fusing sequences encoding for Tat signal peptides in frame to the lignin peroxidase gene.
Figure 7:TAT pathways can export entire folded proteins, while Sec involves an unfolding step, export of the unfolded protein and a refolding of the unfolded protein. Both pathways result in a folded protein in the periplasm.
Another difficulty is the crucial addition of the heme group. Producing lignin peroxidase will only give us nonfunctional polypeptides if we do not have enough heme. We decided to focus up regulation on aminolevulinic acid (ALA) synthesis since it is the rate limiting step for heme production. However, instead of the time consuming up regulation of enzymes responsible for ALA, we decided to simply add ALA to the media, which would then be taken up by e coli. This chemical approach cuts out the rate limiting step entirely and is a cheap method that is realistic for the short time scale of iGEM research.
Sorbitol Biosynthesis
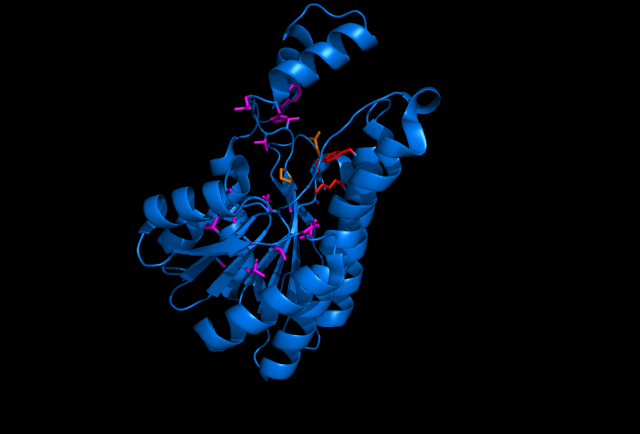
Sorbitol anabolism is part of the glycolysis pathway in E.coli. In the pathway, interconversion between fructose and sorbitol is catalyzed by sorbitol dehydrogenase(SDH), a monomeric enzyme that uses NADH in the process. Natural expression of SDH is generally up-regulated only in the presence of sorbitol as a carbon source. The initial idea to stimulate sorbitol over-production was to simply insert a plasmid containing the gene which codes for SDH and grow the cells in an environment lacking sorbitol thus pushing the metabolic reaction governing fructose and sorbitol interconversion towards sorbitol.
Sorbitol Dehydrogenase NADH binding site (purple wire frame), sorbitol active site (red wire frame), sorbitol hydrogen bonding amino acids (orange wire frame, doesn't include hydrogen bonding from the sorbitol active site) This image was made with PDB file 1K2W on pymol.
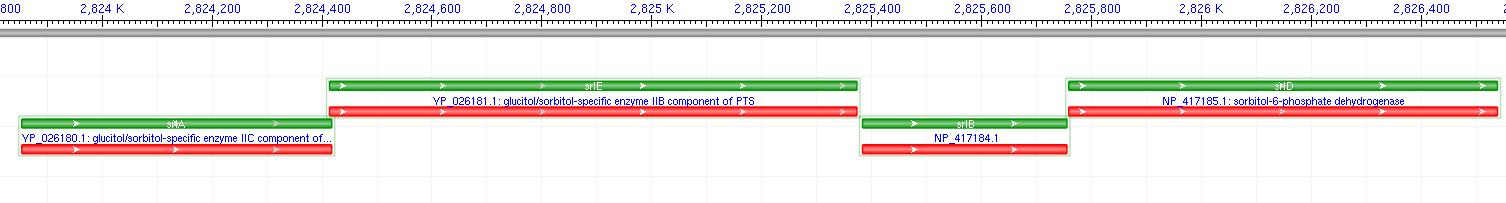
The gene encoding for SDH, srlD, is part of the srl operon in E.coli (operon pictured below). Information on the SDH assembly pathway and enzymatic activity is limited. Consequently we decided that insertion of both the srlD gene and srl operon into vectors would take more time initially but would potentially be beneficial due to the chance that the gene alone would not result in effective SDH production.
Figure 6: SRL Operon
Increased cellular production of SDH would most likely increase the amount of sorbitol produced by the cell, however it could not alone account for the type of mass production that would make sorbitol biosynthesis commercially useful. In order to determine other methods of increasing sorbitol production, our group turned to computer modeling.
Phosphofructokinase Knockout
Figure 7: PFK Knockout in the metabolic pathway.
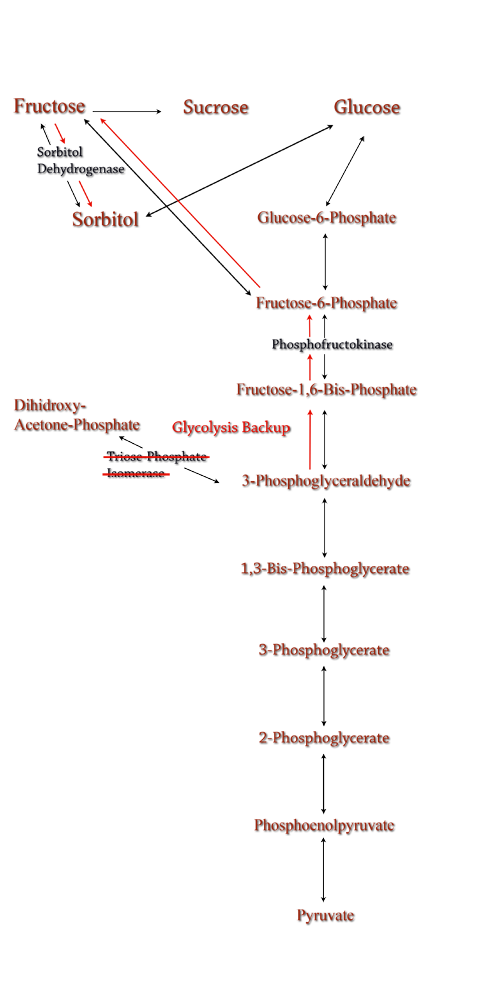
The idea behind a phosphofructokinase (PFK) knockout is to literally stop the glycolysis pathway after fructose-6-phosphate. The backup of metabolic products caused by the knockout should pool and form a favorable pathway for increased sorbitol production. The obvious problem with this procedure is that knocking out a part of the glycolysis pathway under normal circumstances kills the cell. This is also why modeling didn't prove useful as the program showed the cell would be dead. We set out to determine how to bypass this normal cellular function in order to use it to increase sorbitol production.
Experiments have been successfully done to create a viable PFK chromosomal knockouts that contain inducible plasmids encoded with the pfk gene. After obtaining this strain we have done countless growth curves with varying media to create ideal conditions for cell growth. After the optimal media is obtained, the addition of a plasmid containing srlD or the srl operon will be introduced to create what we hope will be this metabolic pathway(right).
Triose Phosphate Isomerase Knockout
Figure 8: TPI Knockout in the metabolic pathway.
Computer modeling was done to determine gene knockouts that increased cellular flux towards sorbitol production. The model was given to us by Dr. Jennie Reed of University of Wisconsin-Madison. More extensive information about modeling can be found at our modeling page. Through the use of our model we determined that knocking out the gene encoding for triose phosphate isomerase would be the most effective at increasing sorbitol production when combined with srl gene up-regulation.
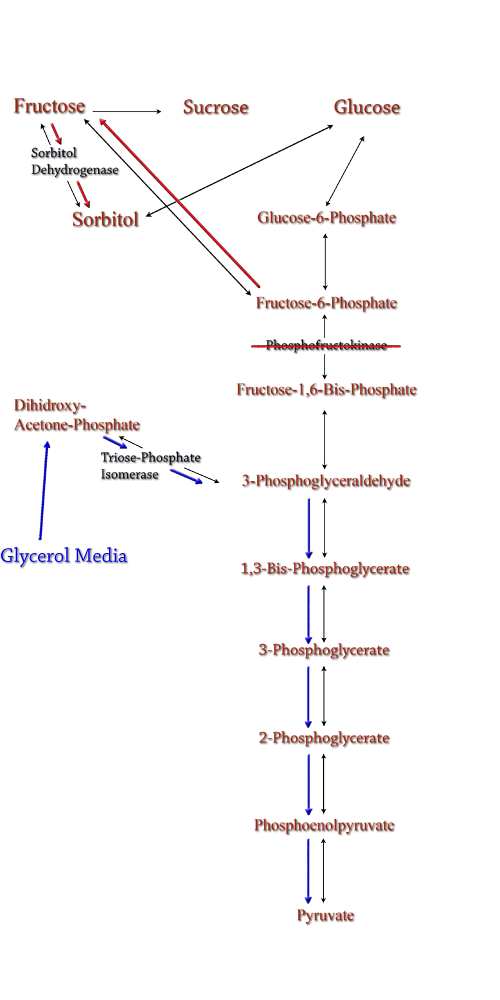
Triose phosphate isomerase(TPI) is another enzyme in E.coli's glycolysis pathway. Its function is to catalyze the interconversion of 3-Phosphoglyceraldehyde to Dihidroxy-acetone-phosphate and back. Knocking out the gene which encodes TPI stops this interconversion, a process which removes products of glycolysis from the cellular reaction mixture shifting the overall reaction downwards towards pyruvate. In essence, knocking out the TPI gene backs up glycolysis and produces a shift towards sorbitol (Pictured right).
TPI is a nonessential gene and as such we were able to obtain it from the Keio collection, a collection of non-essential single gene knockouts from the E.coli K-12 derivative strain BW25113. The increased cellular flux modeled in a TPI knockout combined with increased intracellular SDH production from a plasmid containing either the srlD gene or srl operon will hopefully result in a large scale production of cellular sorbitol.
Even with the promising modeling results, our group worked on alternative routes for increased cellular sorbitol. One of these routes, knocking out phosphofructokinase, was an additional project that our group undertook.
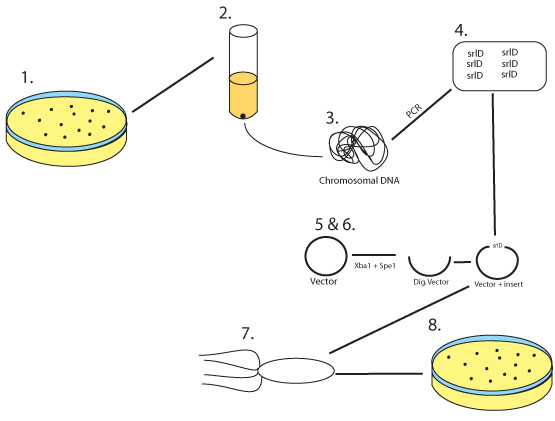
Approach
Figure 9: 1. Wild Type E.coli MG1655 is grown on an LB plate
2. A colony is selected and grown overnight
3. The chromosomal DNA is harvested from the over night culture
4. srlD gene is cloned and amplified via a PCR reaction adding restriction sites for Xba1 and Spe1 at either end
5. The biobrick vector pSB1AK3 as well as the srlD PCR product are digested with Xba1 and Spe1
6. The two digestion products are then ligated together
7. The ligation product is then transformed into competent E.coli
8. E. coli is then plated on LB plates containing Amp/Kan to select for transformants
References
Guzman L., Belin D., CarsonM., and Beckwith L. (1995). Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. Journal of Bacteriology. 177(14): 4121-4130.
Lorenzo Nissen, Gaspar Pérez-Martínez, María J. Yebra (2005) Sorbitol synthesis by an engineered Lactobacillus casei strain expressing a sorbitol-6-phosphate dehydrogenase gene within the lactose operon �FEMS Microbiology Letters 249 (1) , 177–183 doi:10.1016/j.femsle.2005.06.010
Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222.
Ladero, V., Ramos, A., Wiersma, A., Goffin, P., Schanck, A., Kleerebezem, M., et al. (2007) High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering. Appl Environ Microbiol 73: 1864–1872.
Metzger J (2006) Production of liquid hydrocarbons from biomass. Angew. Chem. Int. Ed. 2006, 45, 696-698.
Lovingshimera, M. R., Siegeleb, D., & Reinharta, G. D. (2006). Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources. Protein Expression and Purification , 46 (2), 475-482.
Karacaoğlan, V., & Özer, I. (2005). Steady-state kinetic properties of sorbitol dehydrogenase from chicken liver. Comparative Biochemistry and Physiology , 140, 309-312.
Archibald, F. S. "A new assay for lignin-type peroxidases employing the dye azure B." Applied and Environmental Microbiology, 58(9), 3110-3116.
"New Method Rapidly Produces Low-Cost Biofuels From Wood, Grass". Energy Daily (2008-04-14).
The International Energy Agency. 26 Aug. 2008. Biofuels for Transport: An International Perspective. April 2004. The International Energy Agency. 26 Aug. 2008. <http://www.iea.org/textbase/nppdf/free/2004/biofuels2004.pdf>
Doyle, Wendy A., Andrew T. Smith. “Expression of lignin peroxidase H8 in Escherichia coli: folding and activation of the recombinant enzyme with Ca2+ and haem.” Biochemistry Journal 315 (1996): 15-19.
Hammel, Kenneth E., Dan Cullen. "Role of fungal peroxidases in biological ligninolysis." Current Opinion in Plant Biology 11 (2008): 349-355.
"Cellulose." Encyclopedia Britannica. 2008. Encyclopedia Britannica Online. 28 Oct. 2008.
DeLisa, Matthew P., Danielle Tullman, and George Georgiou. "Folding quality in the export of proteins by the bacterial twin arginine translocation pathway." Proceedings of the National Academy of Sciences 100 (2003): 6155-6120.
Tullman-Ereck, Danielle, Matthew P. DeLisa, Yasuaki Kawarasaki, Pooya Iranpour, Brian Ribnicky, Tracy Palmer, and George Georgiou. "Export Pathway Selectivity of Escherichia Coli Twin Arginine Translocation Signal Peptides." Journal of Biological Chemistry 282 (2007): 8309-83116.
Brenda: the Comprehensive Enzyme Information System. 2008.2. Technical University of Braunschweig Department of Bioinformatics and Biochemistry. 2 July 2008. <http://www.brenda-enzymes.info/>
hello
Guzman L., Belin D., CarsonM., and Beckwith L. (1995). Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. Journal of Bacteriology. 177(14): 4121-4130.
Thomas Choinowski, Wolfgang Blodig, Kaspar H. Winterhalter and Klaus piontek (1999) The crystal structure of lignin peroxidase at 1.70A resolution reveals a hydrozy group on the Cb of tryptophan171: A novel radical site formed during the redox cycle. Journal of Molecular Biology 286, 809-827
Thomas L Poulos, Steven L Edwards, Hiroyuki Wariishii, and Michael H Gold (1993) Crystallographic reginement of lignin peroxidase at 2A. The journal of biological chemistry 268, 4429-4440
Wendy A Doyel, Wolfgang Blodig, Nigel C Veitch, Klaus Piontek and Andrew T smith (1998) Two substrate intereaction sites in lignin peroxidas e revealed by site directed mutagenesis. Biochemistry 37, 15097-15105
 "
"