Team:Heidelberg/Notebook/Killing II/9thweek
From 2008.igem.org
(→Receiver-Colicin E9/E9_lys) |
(→Sunday10/05/2008) |
||
| (16 intermediate revisions not shown) | |||
| Line 559: | Line 559: | ||
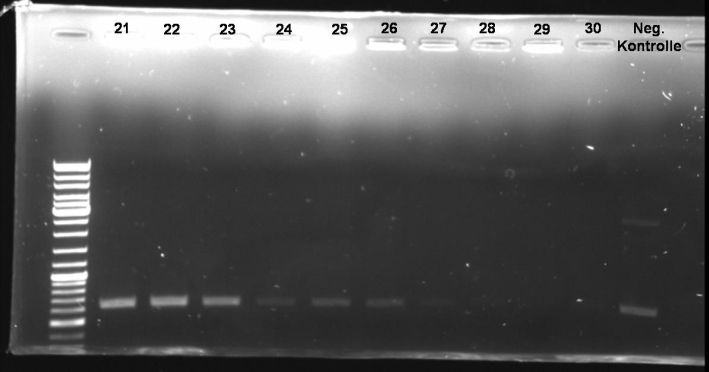
*Gel of Digestion: 1% Agarose, 135 V, 30 min [[Image:080929_digestion_of_ColE9-lys_pSB1A3_receiver_cloning_small.jpg |500 px | center]] | *Gel of Digestion: 1% Agarose, 135 V, 30 min [[Image:080929_digestion_of_ColE9-lys_pSB1A3_receiver_cloning_small.jpg |500 px | center]] | ||
| - | *Results of Digestion: | + | *Results of Digestion: |
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | ||
==Tuesday 09/30/2008== | ==Tuesday 09/30/2008== | ||
| Line 590: | Line 591: | ||
*Plan for activity tests of Colicin-Receivers: | *Plan for activity tests of Colicin-Receivers: | ||
**Overnight measurement (Wednesday - Thursday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE9/ColE9lys cells, supernatant and lysed supernatant. | **Overnight measurement (Wednesday - Thursday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE9/ColE9lys cells, supernatant and lysed supernatant. | ||
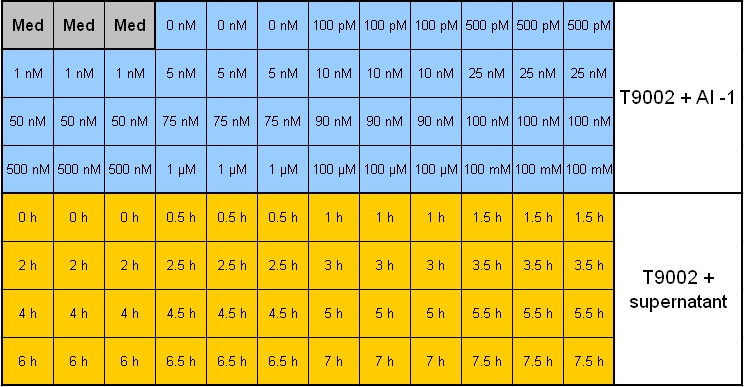
| - | **Pipetting scheme: [[Image:080930_pipetting_scheme_01.jpg | | + | **Pipetting scheme: [[Image:080930_pipetting_scheme_01.jpg |800 px | center]] |
| - | **Legend_1: [[Image:080930_pipetting_scheme_legend_1.jpg | | + | **Legend_1: [[Image:080930_pipetting_scheme_legend_1.jpg |180 px | center]] |
**Legend_2: [[Image:080930_pipetting_scheme_legend_2_small.jpg |500 px | center]] | **Legend_2: [[Image:080930_pipetting_scheme_legend_2_small.jpg |500 px | center]] | ||
**Inocculation of Overnightcultures: | **Inocculation of Overnightcultures: | ||
| Line 633: | Line 634: | ||
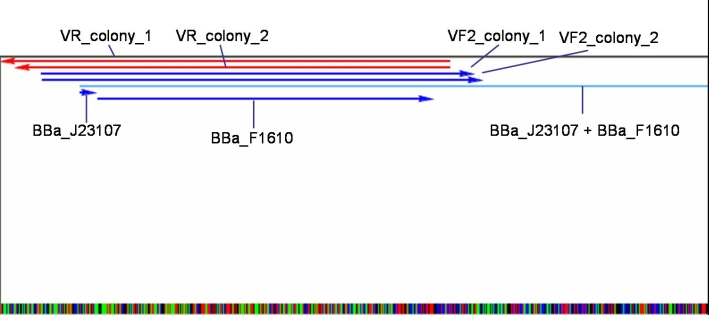
*Sequencing results: Both colonies contain the right fragment. So we inocculate new overnight cultures for Minipreps, Glycerolstocks and activity tests. [[Image:080930_J23107_BBaF1610_contig_small.jpg |500 px | center]] | *Sequencing results: Both colonies contain the right fragment. So we inocculate new overnight cultures for Minipreps, Glycerolstocks and activity tests. [[Image:080930_J23107_BBaF1610_contig_small.jpg |500 px | center]] | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | |
| - | + | ||
| - | + | ||
| - | + | ||
==Wednesday 10/01/2008== | ==Wednesday 10/01/2008== | ||
| Line 643: | Line 641: | ||
*Gel of pSB1A3-Receiver-Col E1 PCR-Singlescreen: 1% Agarose, 135 V, 30 min | *Gel of pSB1A3-Receiver-Col E1 PCR-Singlescreen: 1% Agarose, 135 V, 30 min | ||
*Gel result: | *Gel result: | ||
| - | + | [[Image: 081001_colE1_singlescreening.jpg | 500 ox | center]] | |
| - | + | Primer Col E1 (above): | |
| - | + | - T9002_LuxpR_Eco_Xba_G_fw | |
| - | + | - ColE1_prot_rv_A_SpeI | |
| - | + | ||
| + | Expected Fragment: | ||
| + | ~ 3200 bp | ||
| + | |||
| + | Primer Col E1 His (below): | ||
| + | - T9002_LuxpR_Eco_ Xba_G_fw | ||
| + | - ColE1_kil_rv_HindIII | ||
| + | |||
| + | Expected Fragment: | ||
| + | ~ 2150 bp | ||
| + | |||
| + | Comment: | ||
| + | Possible explanation for the high amount of bands. | ||
| + | Maybe annealing temperature was to low and primers annealed anywhere. | ||
| + | -> Sequencing for colony 6, 38 & 39 | ||
| + | |||
| + | |||
<br> | <br> | ||
*Digestion of PCR-product Colicin E9, E9lys with right prefix and sufix: 2h 40 min at 37 °C | *Digestion of PCR-product Colicin E9, E9lys with right prefix and sufix: 2h 40 min at 37 °C | ||
| Line 660: | Line 674: | ||
**Gel of pSB1A3-Receiver-Col E9/E9 lys digestion: 1% Agarose, 135 V, 30 min | **Gel of pSB1A3-Receiver-Col E9/E9 lys digestion: 1% Agarose, 135 V, 30 min | ||
*Gel result: | *Gel result: | ||
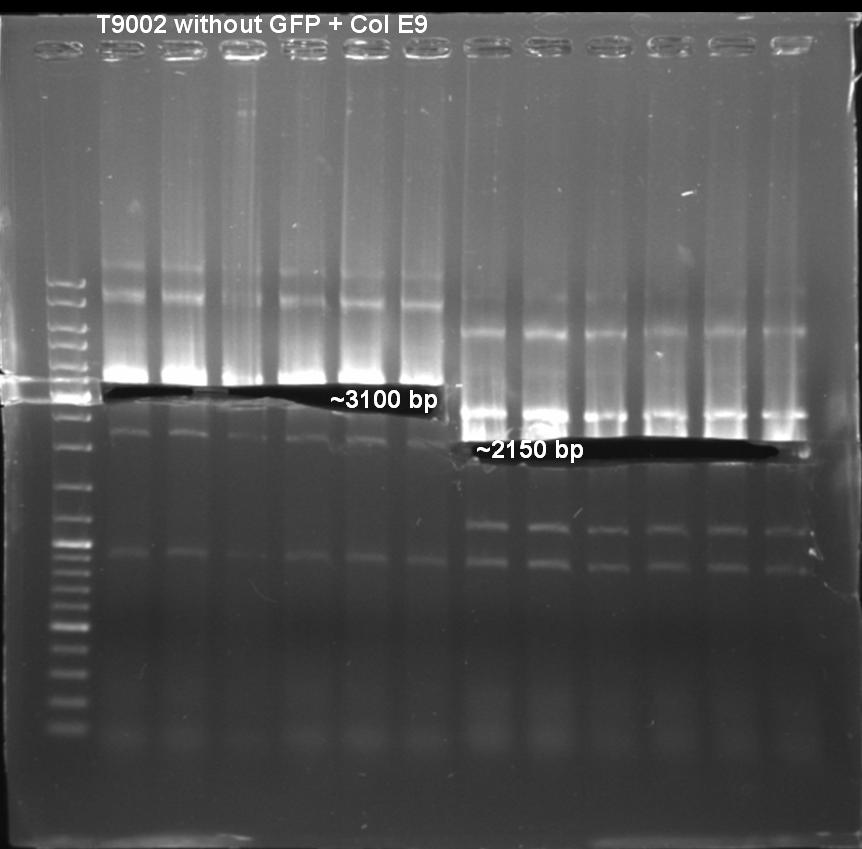
| - | + | [[Image: 081001_colE9_colE9lys_BB.jpg |500 px | center]] | |
| - | + | ||
| - | + | Primer Col E9: | |
| - | + | - T9002_LuxpR_Eco_ Xba_G_fw | |
| - | + | - ColE9_plasmid_rv_A_SpeI | |
| + | |||
| + | Expacted Fragment: | ||
| + | ~ 3100 bp | ||
| + | |||
| + | Primer Col E9 lys: | ||
| + | - T9002_LuxpR_Eco_ Xba_G_fw | ||
| + | - ColE9_lysProt_rv_A_SpeI | ||
| + | |||
| + | Expacted Fragment: | ||
| + | ~ 2150 bp | ||
| + | |||
| + | Comment: | ||
| + | Possible explanation for the high amount of bands. | ||
| + | May be annealing temperature was to low and primers annealed anywhere. | ||
| + | |||
<br> | <br> | ||
*Digestion of : 2h 40 min at 37 °C | *Digestion of : 2h 40 min at 37 °C | ||
| Line 678: | Line 707: | ||
*Gelextraction (?) | *Gelextraction (?) | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Thursday 10/02/2008== | ==Thursday 10/02/2008== | ||
| Line 708: | Line 731: | ||
14 h 16°C --> 20 min 65°C | 14 h 16°C --> 20 min 65°C | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | ||
| - | + | ==Friday 10/03/2008== | |
| - | + | ===pSB1A2-Receiver-Colicin cloning=== | |
| - | + | Transformation of the ON ligations of yesterday. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | |
===Colicin activity test=== | ===Colicin activity test=== | ||
| + | *Different colicin producing cells were sonified: | ||
| + | **naturally colicin producing cells (colE1 and colE9) stressed with mitomycin C and unstressed | ||
| + | **cells containing different colicin-receiver cloning constructs (colE1, colE9 and colE9lys) | ||
| + | **reference cells (T9002 without GFP) | ||
| + | *this supernatants were mixed with GFP producing prey cells in different ratios | ||
| + | *measurement of OD and GFP over night | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Saturday 10/04/2008== | ==Saturday 10/04/2008== | ||
| Line 761: | Line 771: | ||
===Sender Cloning: constitutive promotor - sender=== | ===Sender Cloning: constitutive promotor - sender=== | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | ||
==Sunday10/05/2008== | ==Sunday10/05/2008== | ||
| Line 822: | Line 833: | ||
**Overnight measurement (Sunday - Monday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE1 cells. | **Overnight measurement (Sunday - Monday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE1 cells. | ||
**Plate scheme (click for detailed picture): [[Image: 081007-pipetting_scheme_sender_test.jpg | 700 px | center ]] | **Plate scheme (click for detailed picture): [[Image: 081007-pipetting_scheme_sender_test.jpg | 700 px | center ]] | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/9thweek back]] | ||
| + | |||
[[Team:Heidelberg/Notebook/Killing_II/10thweek | go to 10<sup>th</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/10thweek | go to 10<sup>th</sup> week]] | ||
Latest revision as of 21:41, 28 October 2008


9th week
Contents |
Monday 09/29/2008
pSB1A3-Receiver-Colicin cloning
- Digestion of pSB1A3-Receiver-ColicinE9/E9lys: E9 -> 61, 63; E9lys -> 34, 35, 47
10 µl DNA 5 µl BSA 10x (NEB) 5 µl EcoRI Buffer (NEB) 1 µl EcoRI 1 µl PstI 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Results of Digestion:
HisTag cloning of Colicins for purification
- PCR-Screen of pQE-30-Col E9 His
25.0 µl Taq Master Mix (Fermentas) 2.5 µl ColE9_prot_fw_BamHI 2.5 µl ColE9_prot_rv_XmaI 20.0 µl H2O 1 colony ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
- Gel of E9 PCR-Screen: 1% Agarose, 135 V, 30 min
- Gel-Results of E9-PCR-Screen: Negative controll shows right band size -> its maybe a supercoiled form of the plasmid. No positive clones.
Sender cloning: pBAD-sender
- Digestion of Sender pBAD: 1h 40min at 37°C
10 µl DNA (1) 5 µl NEBuffer 2 (NEB) 5 µl BSA 10x 1 µl XbaI 1 µl SpeI 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Results of Digestion:
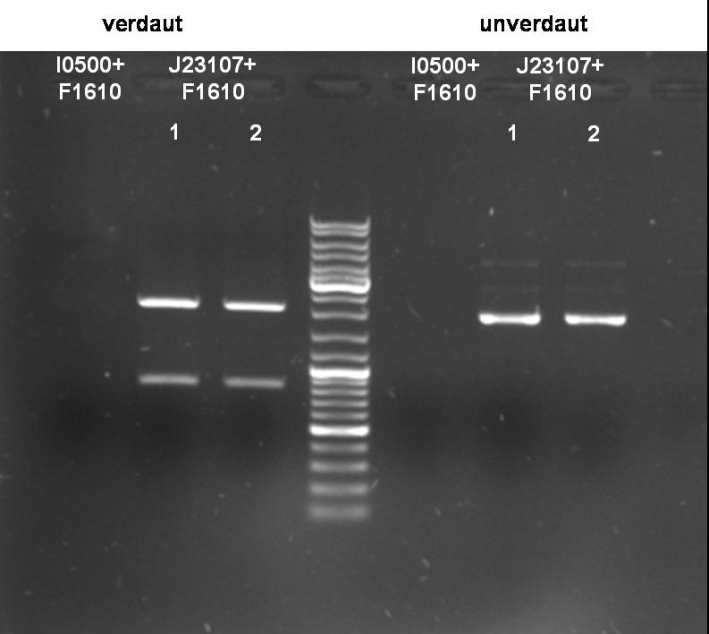
- I0500+F1610:
- Expected Fragments:
- ~ 2030bp (Insert: I0500+F1610)
- ~ 4425bp (Backbone: pSB2K3)
- Expected Fragments:
- I0500+F1610:
No bands visible. Probably no DNA or low concentrated DNA. Nanodrop measurement ~80ng/µl. Despite of neg. digestion we sended one probe to GATC for sequencing.
- J23107+F1610:
- Expected Fragments:
- ~ 850bp (Insert: J23107+F1610)
- ~ 2070bp (Backbone: J61002)
- Expected Fragments:
- J23107+F1610:
Expected bands are visible on the gel. Probes send for sequencing (GATC.
Sender cloning: constitutive promotor-sender
- Digestion of Sender with constitutive promotor: 1h 40min at 37°C
10 µl DNA (1) 5 µl NEBuffer 2 (NEB) 5 µl BSA 10x 1 µl XbaI 1 µl SpeI 28 µl H2O ----- 50 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Results of Digestion:
[back]
Tuesday 09/30/2008
pSB1A3-Receiver-Colicin cloning
Receiver-Colicin E9/E9_lys
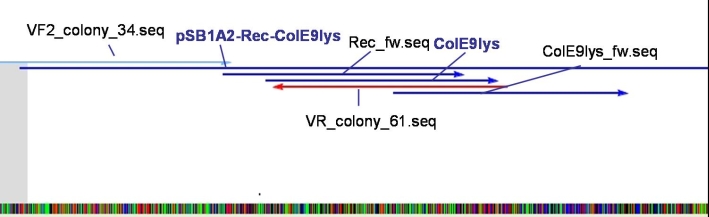
- Sequencing results: The colonies 34, 35 and 47 contains the right insert. But the sequences behind the BioBrick Suffix do not fit to [http://partsregistry.org/Part:pSB1A3 pSB1A3] vector. The sequence which we received from GATC has 78 bp less than the one from partsregistry.org. To fix this problem we aligned our sequencing results against the pSB1A2 vector and they fit perfect. Because of that we assume that our BBa_T9002 is inside the pSB1A2 vector and not as expected in the pSB1A3 vector.
- Sequencing results: The colonies 61 and 63 contains the right insert, but like ColE9lys in [http://partsregistry.org/Part:pSB1A2 pSB1A2] vector.
- PCR for amplification of Receiver-Colicin E9/E9_lys for recloning with the right prefix and suffix
25.0 µl Phusion Master Mix (Finnzymes, NEB) 2.5 µl T9002_LuxpR_Not_Eco_Xba_G_fw (Tm=80,96°C) 2.5 µl ColE9_plasmid_rv_A_SpeI(Tm=57,48°C)/ColE9_lysProt_rv_A_SpeI (Tm=64,29°C) 20.0 µl H2O 2.0 µl Miniprep (E9: Colony 61, E9lys: Colony 34) ------- 50.0 µl
program:
95 °C 30 sec 95 °C 10 sec | 54 °C 30 sec | 25 cycles 72 °C 1 min 30 sec | 72 °C 10 min 4 °C constant
- Gel of E9 and E9 lys PCR-amplification: 1% Agarose, 135 V, 30 min
- Gel-Results of E9-PCR-amplification:
- Gel-Results of E9lys-PCR-amplification:
- Plan for activity tests of Colicin-Receivers:
- Overnight measurement (Wednesday - Thursday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE9/ColE9lys cells, supernatant and lysed supernatant.
- Pipetting scheme:
- Legend_1:
- Legend_2:
- Inocculation of Overnightcultures:
- 10 ml M9 + Kanamycin media + Reference promoter cells (BBa_I20260)
- 6 x 3 ml M9 Media + pSB1A3-Receiver without GFP
- 6 x 3 ml M9 Media + pSB1A3-Receiver-ColE9
- 6 x 3 ml M9 Media + pSB1A3-Receiver-ColE9lys
Receiver-Colicin E1
- PCR-Screen of pSB1A3-Receiver-Colicin E1 Cloning
25.0 µl Taq Master Mix (Fermentas) 2.5 µl ColE1_prot_fw_BamHI 2.5 µl ColE1_prot_rv_HindIII 20.0 µl H2O 5 colonies ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 54 °C 1 min | 30 cycles 72 °C 1 min | 72 °C 10 min 4 °C constant
- Gel of Col E1 PCR-Screen: 1% Agarose, 135 V, 30 min
- Gel-Results of E1-PCR-Screen:
- Expected Fragmentsize: ~1580 bp
- Colonies 6-10 and 36-40 show the right fragment size. -> New colony-PCR-Screen.
HisTag cloning of Colicins for purification
Sender cloning: pBAD-sender
- Sequencing results: No sequence -> Inocculation of new ONC
Sender cloning: constitutive promotor-sender
- Sequencing results: Both colonies contain the right fragment. So we inocculate new overnight cultures for Minipreps, Glycerolstocks and activity tests.
[back]
Wednesday 10/01/2008
pSB1A3-Receiver-Colicin cloning
- Gel of pSB1A3-Receiver-Col E1 PCR-Singlescreen: 1% Agarose, 135 V, 30 min
- Gel result:
Primer Col E1 (above): - T9002_LuxpR_Eco_Xba_G_fw - ColE1_prot_rv_A_SpeI
Expected Fragment: ~ 3200 bp
Primer Col E1 His (below): - T9002_LuxpR_Eco_ Xba_G_fw - ColE1_kil_rv_HindIII
Expected Fragment: ~ 2150 bp
Comment: Possible explanation for the high amount of bands. Maybe annealing temperature was to low and primers annealed anywhere. -> Sequencing for colony 6, 38 & 39
- Digestion of PCR-product Colicin E9, E9lys with right prefix and sufix: 2h 40 min at 37 °C
38,5 µl DNA (PCR-amplification) 1 µl SpeI (NEB) 0,5 µl EcoRI (NEB) 5 µl NEBuffer EcoRI (NEB) 5 µl BSA 10x (NEB) ----- 50 µl
- Gel of pSB1A3-Receiver-Col E9/E9 lys digestion: 1% Agarose, 135 V, 30 min
- Gel result:
Primer Col E9: - T9002_LuxpR_Eco_ Xba_G_fw - ColE9_plasmid_rv_A_SpeI
Expacted Fragment: ~ 3100 bp
Primer Col E9 lys: - T9002_LuxpR_Eco_ Xba_G_fw - ColE9_lysProt_rv_A_SpeI
Expacted Fragment: ~ 2150 bp
Comment: Possible explanation for the high amount of bands. May be annealing temperature was to low and primers annealed anywhere.
- Digestion of : 2h 40 min at 37 °C
9 µl DNA (Miniprep of ColE9 colonies 61 and 63) 1 µl SpeI (NEB) 0,5 µl EcoRI (NEB) 5 µl NEBuffer EcoRI (NEB) 5 µl BSA 10x (NEB) 28,5 µl H2O ----- 50 µl
- Gelextraction (?)
[back]
Thursday 10/02/2008
pSB1A2-Receiver-Colicin cloning
- By sequencing we figured out, that the plasmid-backbone we used (and are still using) is not the pSB1A3 as it should be but the pSB1A2. Sequencing of the registery part BBa_T9002, where we cut out the plasmid-backbone we are using, affirmed that this part is not on the pSB1A3 plasmid as mentioned but on the pSB1A2. Because we have no working registry part on a pSB1A3 plasmid available in lab, we decided to continue with the pSB1A2 backbone and will also do our standardization with this plasmid.
Sequencing results of T9002/GFP_ColE1 cloning
- two of the sequenced colonies gave positive results: Colony 6 and Colony 39
- --> two positive colE1 cloning constructs: T9002/GFP_ColE1.6 and T9002/GFP_ColE1.39
Continuance of T9002/GFP_colE9 and _colE9lys cloning for standardization (with correct pre-/suffixes)
- gel (0,7 %, 135 V, 60 min) of the restriction digest done yesterday (EcoRI + PstI) with the samples for the backbone (61, 63, 63)
- pSB1A2 at ~2000 bp where cut out
- gel extraction with these bands (61, 63, 63) and PCR purification with the restriction digests done yesterday (EcoRI + PstI) with the samples ColE9 and ColE9lys. An additional made analytical gel of these two samples reminded us, that the colE9 fragment contains an EcoRI site inside the fragment. For this reason, we could not use the digested colE9 fragment and continued the cloning only with the colE9lys fragment.
- overnight ligation of pSB1A2 (9,1 ng/µl) and colE9lys (14,4 ng/µl)
2 µl T4 ligase buffer 10x (Fermentas) 2 µl T4 ligase (Fermentas) 13 µl ColE9lys 3 µl pSB1A2 ----- 20 µl
14 h 16°C --> 20 min 65°C
[back]
Friday 10/03/2008
pSB1A2-Receiver-Colicin cloning
Transformation of the ON ligations of yesterday.
Colicin activity test
- Different colicin producing cells were sonified:
- naturally colicin producing cells (colE1 and colE9) stressed with mitomycin C and unstressed
- cells containing different colicin-receiver cloning constructs (colE1, colE9 and colE9lys)
- reference cells (T9002 without GFP)
- this supernatants were mixed with GFP producing prey cells in different ratios
- measurement of OD and GFP over night
[back]
Saturday 10/04/2008
pSB1A3-Receiver-Colicin cloning
- Inocculation of Overnightcultures:
- 25 ml TB + Kanamycin media + Reference promoter cells (BBa_I20260)
- 25 ml TB + Ampicillin media + pSB1A3-Receiver without GFP
- 25 ml TB + Ampicillin media + pSB1A3-Receiver-ColE1
- Inocculation of Overnightcultures:
HisTag cloning of Colicins for purification
Sender Cloning: pBAD - sender
Sender Cloning: constitutive promotor - sender
[back]
Sunday10/05/2008
pSB1A3-Receiver-Colicin cloning
Colicin E9 lys
- PCR-Screen of Colonies (Transformation from Fr,10/03/2008) of pSB1A3-Receiver-Col E9 lys
25.0 µl Taq Master Mix (Fermentas) 2.5 µl T9002_LuxpR_Not_Eco_Xba_G_fw (Tm=80,96°C) 2.5 µl ColE9_lyProt_rv_A_SpeI (Tm=64,29°C) 20.0 µl H2O 1 colony ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 64 °C 1 min | 30 cycles 72 °C 1 min 30 sec | 72 °C 10 min 4 °C constant
- Inocculation of Overnightcultures:
- 12 ml TB + Kanamycin media + Reference promoter cells (BBa_I20260)
- 23 x 2 ml TB + Ampicillin media + pSB1A3-Receiver without GFP
- each 2 ml induced with the following AI-1 concentration:
- Inocculation of Overnightcultures:
0 nM 200 nM 60 nM 250 nM 70 nM 300 nM 80 nM 350 nM 90 nM 400 nM 100 nM 500 nM 110 nM 600 nM 120 nM 700 nM 130 nM 800 nM 140 nM 900 nM 150 nM 1 µM 160 nM
- 23 x 2 ml TB + Ampicillin media + pSB1A3-Receiver-ColE9 lys
- each 2 ml induced with the following AI-1 concentration:
- 23 x 2 ml TB + Ampicillin media + pSB1A3-Receiver-ColE9 lys
0 nM 200 nM 60 nM 250 nM 70 nM 300 nM 80 nM 350 nM 90 nM 400 nM 100 nM 500 nM 110 nM 600 nM 120 nM 700 nM 130 nM 800 nM 140 nM 900 nM 150 nM 1 µM 160 nM
Colicin E1
- Plan for activity tests of Colicin-Receivers:
- Overnight measurement (Sunday - Monday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A3-Rec-ColE1 cells.
- Plate scheme (click for detailed picture):
[back]
 "
"