October
From 2008.igem.org
(Difference between revisions)
m |
|||
| (38 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
Content= | Content= | ||
<div style="font-size:18pt;"> | <div style="font-size:18pt;"> | ||
| - | <font face="Arial Rounded MT Bold" style="color:#010369"> | + | <font face="Arial Rounded MT Bold" style="color:#010369">_october</font></div> |
<br> | <br> | ||
<br> | <br> | ||
| Line 97: | Line 97: | ||
<br> | <br> | ||
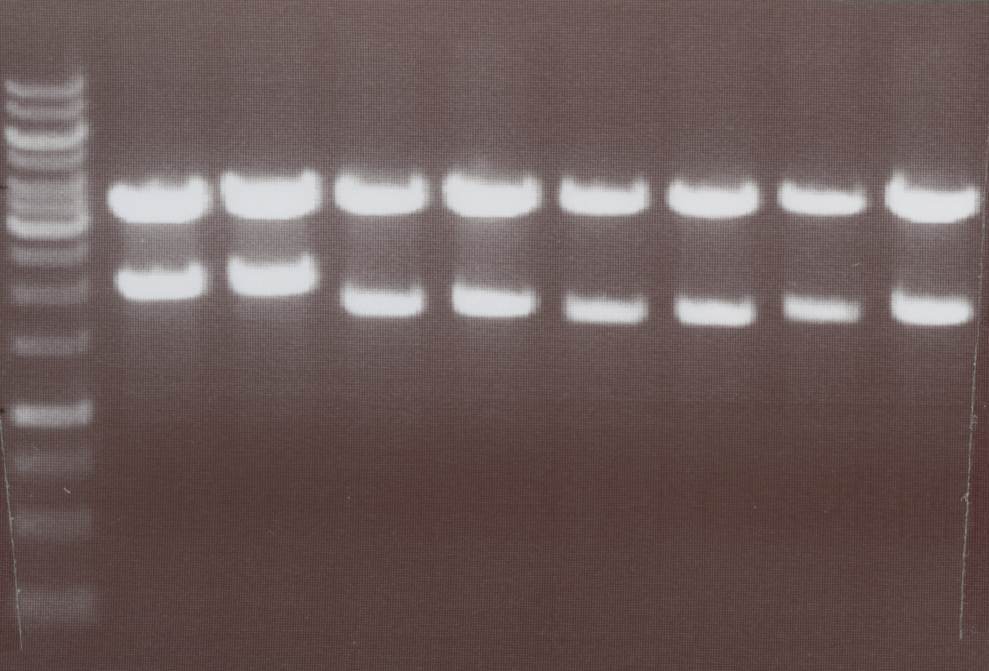
'''Analytic digestion of:''' (Sabine)<br> | '''Analytic digestion of:''' (Sabine)<br> | ||
| - | -pMA-splitlinker-Cerulan split C-CFP -> | + | -pMA-splitlinker-Cerulan split C-CFP -> expected bands: pMA-Vector ~ 2360bp, splitlinker-cCFP ~ 370bp<br> |
| - | -pMA-splitlinker-Venus split C-YFP -> | + | -pMA-splitlinker-Venus split C-YFP -> expected bands: pMA-Vector ~ 2360bp, splitlinker-cYFP ~ 370bp<br> |
| - | [[Image:TeamFreiburg2008_Linker+cCFP_1005.jpg|250px]] | + | [[Image:TeamFreiburg2008_Linker+cCFP_1005.jpg|250px]]<br> |
| - | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | |
| + | Lane02: pMA-splitlinker-Cerulan split C-CFP clone 1 (correct)<br> | ||
| + | Lane03: pMA-splitlinker-Cerulan split C-CFP clone 2 (not correct)<br> | ||
| + | Lane04: pMA-splitlinker-Cerulan split C-CFP clone 3 (correct)<br> | ||
| + | Lane05: pMA-splitlinker-Cerulan split C-CFP clone 4 (correct)<br> | ||
| + | Lane06: ---<br> | ||
| + | Lane07: pMA-splitlinker-Venus split C-YFP clone 1 (correct)<br> | ||
| + | Lane08: pMA-splitlinker-Venus split C-YFP clone 2 (not correct)<br> | ||
| + | Lane09: pMA-splitlinker-Venus split C-YFP clone 3 (not correct)<br> | ||
| + | Lane10: pMA-splitlinker-Venus split C-YFP clone 4 (correct)<br> | ||
<br> | <br> | ||
'''Preparative digestion''' (Sabine)<br> | '''Preparative digestion''' (Sabine)<br> | ||
| Line 130: | Line 139: | ||
-pMA-splitlinker-Cerulan split C-CFP<br> | -pMA-splitlinker-Cerulan split C-CFP<br> | ||
-pMA-splitlinker-Venus split C-YFP<br> | -pMA-splitlinker-Venus split C-YFP<br> | ||
| - | <br> | + | <br><br>'''Detection of linkage between Fab and NIP'''(Normann)<br> |
| + | [[Image:Freiburg08Ta50Ringer50_kont+nip.jpg]]<br> | ||
| + | ''Fig: Oriogamis with (left) and without NIP given to B-cells in 50% TA-buffer(12,5mM Mg2+) and 50% Ringer (12,5mM Mg2+) ''<br><br>[[Image:Freiburg08NipnoNipinTa.jpg]]<br> | ||
| + | ''Fig: Origami without (left) and with (right) NIP given to B-cells in TA-buffer (12,5mM Mg2+)''<br> Origamis seem to be absorbed by the cells. A specific linkage was not apparent.Sole TA buffer is osmotically disadvantageous for the cells. | ||
'''Analytic digestion of''' (Kathrin)<br> | '''Analytic digestion of''' (Kathrin)<br> | ||
-transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones<br> | -transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones<br> | ||
| Line 153: | Line 165: | ||
- transfectionvector-CMV PCR product-YFP clones 1-4<br> | - transfectionvector-CMV PCR product-YFP clones 1-4<br> | ||
- transfectionvector-CMV PCR product-Lipocalin clones 5-7<br> | - transfectionvector-CMV PCR product-Lipocalin clones 5-7<br> | ||
| - | + | ||
test digestion of the clones named above with NotI<br> | test digestion of the clones named above with NotI<br> | ||
analysis on the agarosegel shows not the expected bands | analysis on the agarosegel shows not the expected bands | ||
| + | <br><br> | ||
| + | '''Transfection of 293t with tv-cmv-yfp'''<br> | ||
| + | was done in a 6well plate <br> | ||
<br> | <br> | ||
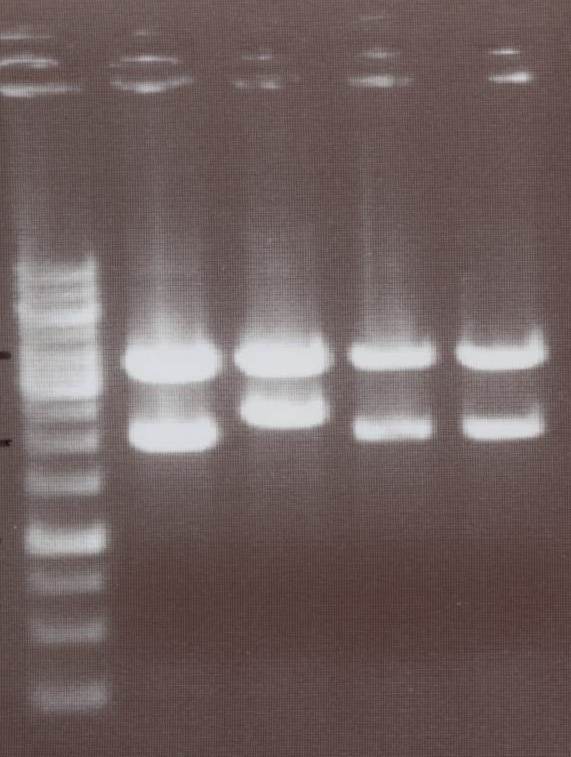
'''Miniprep and analytic digestion''' (Sabine)<br> | '''Miniprep and analytic digestion''' (Sabine)<br> | ||
| - | -pMA-transmembrane | + | 1) pMA-transmembrane-Cerulan N-CFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-Cerulan N-CFP ~ 610bp<br> |
| - | -pMA-transmembrane | + | 2) pMA-transmembrane-Venus N-YFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-Venus N-YFP ~ 610bp<br> |
| - | -pMA-transmembrane+Cerulan N-CFP<br> | + | 3) pMA-transmembrane-splitlinker-Cerulan split C-CFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-splitlinker-Cerulan split C-CFP ~ 430bp<br> |
| - | -pMA-transmembrane | + | [[Image:TeamFreiburg2008_TM+NCFP_NYFP_Li-cCFP_Li-cYFP_1007.jpg|250px]]<br> |
| - | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | |
| - | [[Image:TeamFreiburg2008_TM+NCFP_NYFP_Li-cCFP_Li- | + | Lane02: pMA-transmembrane-Cerulan N-CFP clone 1 (correct)<br> |
| + | Lane03: pMA-transmembrane-Cerulan N-CFP clone 2 (correct)<br> | ||
| + | Lane04: pMA-transmembrane-Cerulan N-CFP clone 3 (correct)<br> | ||
| + | Lane05: pMA-transmembrane-Venus N-YFP clone 1 (correct)<br> | ||
| + | Lane06: pMA-transmembrane-Venus N-YFP clone 2 (correct)<br> | ||
| + | Lane07: pMA-transmembrane-Venus N-YFP clone 3 (correct)<br> | ||
| + | Lane08: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 1 (correct)<br> | ||
| + | Lane09: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 2 (correct)<br> | ||
| + | Lane10: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 3 (correct)<br> | ||
| + | <br> | ||
| + | 4) pMA-transmembrane-splitlinker-Venus split C-YFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-splitlinker-Venus split C-YFP ~ 430bp<br> | ||
| + | [[Image:TeamFreiburg2008_TM+NCFP_NYFP_Li-cCFP_Li-cYFP2_1007.jpg|100px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: pMA-transmembrane-splitlinker-Venus split C-YFP clone 1 (correct)<br> | ||
| + | Lane03: pMA-transmembrane-splitlinker-Venus split C-YFP clone 2 (correct)<br> | ||
| + | Lane04: pMA-transmembrane-splitlinker-Venus split C-YFP clone 3 (correct)<br> | ||
<br> | <br> | ||
| - | |||
| - | |||
<h3>Oct. 8th 2008</h3> <br> | <h3>Oct. 8th 2008</h3> <br> | ||
'''Digestion of CFP''' normann<br> | '''Digestion of CFP''' normann<br> | ||
| - | In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit | + | In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit carrying CFP was digested using pst1 and Xba1. <br> |
The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"<br> | The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"<br> | ||
<br> | <br> | ||
| Line 217: | Line 245: | ||
<br> | <br> | ||
'''Analytic Digestion''' (Sabine)<br> | '''Analytic Digestion''' (Sabine)<br> | ||
| - | -pMA-signalpeptide-Anti-Nip-sc-Fv | + | -pMA-signalpeptide-Anti-Nip-sc-Fv -> expected bands: pMA-Vector ~2360bp, signalpeptide-Anti-Nip-sc-Fv ~880bp <br> |
| - | -pMA-signalpeptide-Lipocalin | + | -pMA-signalpeptide-Lipocalin -> expected bands: pMA-Vector ~2360bp, signalpeptide-Lipocalin ~670bp<br> |
| + | [[Image:TeamFreiburg2008_pMA-SP+NIP+Lipo_1009.jpg|200px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: pMA-signalpeptide-Anti-Nip-sc-Fv clone 1 (correct)<br> | ||
| + | Lane03: pMA-signalpeptide-Anti-Nip-sc-Fv clone 2 (correct)<br> | ||
| + | Lane04: pMA-signalpeptide-Anti-Nip-sc-Fv clone 3 (correct)<br> | ||
| + | Lane05: pMA-signalpeptide-Lipocalin clone 1 (correct)<br> | ||
| + | Lane06: pMA-signalpeptide-Lipocalin clone 2 (correct)<br> | ||
| + | Lane07: pMA-signalpeptide-Lipocalin clone 3 (correct)<br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
'''Preparative Digestion''' (Sabine)<br> | '''Preparative Digestion''' (Sabine)<br> | ||
-pMA-BB057(Splitluciferase) with NgoMIV and SpeI<br> | -pMA-BB057(Splitluciferase) with NgoMIV and SpeI<br> | ||
| Line 289: | Line 322: | ||
'''Analytic Digestion''' (Kathrin)<br> | '''Analytic Digestion''' (Kathrin)<br> | ||
With NotI<br> | With NotI<br> | ||
| - | -pMA-signalpeptide- | + | -pMA-signalpeptide-Anti-Nip-sc-Fv -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Anti-Nip-sc-Fv ~ 880bp<br> |
| + | -pMA-signalpeptide-Anti-Nip-sc-Fv-GGGS-Linker -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Anti-Nip-sc-Fv-GGGSlinker ~ 920bp<br> | ||
| + | -pMA-signalpeptide-Lipocalin -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Lipocalin ~ 670bp<br> | ||
| + | -pMA-signalpeptide-Lipocalin-GGGS-Linker (2nd try) -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Lipocalin-GGGSlinker ~ 710bp<br> | ||
This time pMA-signalpeptide-Lipocalin was also digested to see the different sizes of the constructs with and without GGGS-Linker on the gel<br> | This time pMA-signalpeptide-Lipocalin was also digested to see the different sizes of the constructs with and without GGGS-Linker on the gel<br> | ||
| - | -pMA-signalpeptide-Anti-Nip-sc-Fv | + | [[Image:TeamFreiburg2008_pMA-SP+NIP+GGGS_+lipo+GGGS_1011.jpg|150px]]<br> |
| - | -> pMA-signalpeptide- | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| + | Lane02: pMA-signalpeptide-Anti-Nip-sc-Fv clone 1(correct)<br> | ||
| + | Lane03: pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker clone 1 (not correct)<br> | ||
| + | Lane04: pMA-signalpeptide-Lipocalin clone 1(correct)<br> | ||
| + | Lane05: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 1 (correct)<br> | ||
| + | Lane06: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 2 (correct)<br> | ||
| + | Lane07: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 3 (correct)<br> | ||
<br> | <br> | ||
'''Preparative Digestion''' (Kathrin)<br> | '''Preparative Digestion''' (Kathrin)<br> | ||
| Line 310: | Line 352: | ||
<br> | <br> | ||
<h3>Oct. 13th 2008</h3> <br> | <h3>Oct. 13th 2008</h3> <br> | ||
| + | <br> | ||
| + | '''Transfection of 293t with tv-cmv-yfp''' (Normann) | ||
| + | <br> done in a 6 well plate. No counted seeding this time due to too little cells in the last try.<br> | ||
'''Transformation''' (Kathrin)<br> | '''Transformation''' (Kathrin)<br> | ||
repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1<br> | repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1<br> | ||
| Line 366: | Line 411: | ||
<br> | <br> | ||
<h3>Oct. 14th 2008</h3> <br> | <h3>Oct. 14th 2008</h3> <br> | ||
| + | <h3>!!! New Coding for all parts !!!</h3> cause of to long descriptions<br><br> | ||
| + | '''1. complete parts in pMA-vector'''<br><br> | ||
| + | '''NIP A''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | ||
| + | '''NIP B''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br> | ||
| + | '''NIP C''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br> | ||
| + | '''NIP D''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br> | ||
| + | '''NIP E''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br> | ||
| + | '''NIP F''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | ||
| + | '''NIP G''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | ||
| + | '''NIP H''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> | ||
| + | '''Lipo A''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | ||
| + | '''Lipo B''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br> | ||
| + | '''Lipo C''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br> | ||
| + | '''Lipo D''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br> | ||
| + | '''Lipo E''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br> | ||
| + | '''Lipo F''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | ||
| + | '''Lipo G''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | ||
| + | '''Lipo H''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> | ||
| + | '''2. complete parts in transfection-vector'''<br><br> | ||
| + | '''NIP I''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | ||
| + | '''NIP II''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br> | ||
| + | '''NIP III''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br> | ||
| + | '''NIP IV''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br> | ||
| + | '''NIP V''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br> | ||
| + | '''NIP VI''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | ||
| + | '''NIP VII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | ||
| + | '''NIP VIII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> | ||
| + | |||
| + | '''Lipo I''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | ||
| + | '''Lipo II''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br> | ||
| + | '''Lipo III''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br> | ||
| + | '''Lipo IV''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br> | ||
| + | '''Lipo V''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br> | ||
| + | '''Lipo VI''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | ||
| + | '''Lipo VII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | ||
| + | '''Lipo VIII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> | ||
| + | '''Transfection''' (Normann) <br> | ||
| + | To test the functionality of the TV+CMV construct, a testtransfection with the construct+YFP in 293t-cells was done.<br><br> | ||
'''Miniprep''' (Kathrin)<br> | '''Miniprep''' (Kathrin)<br> | ||
- 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells<br> | - 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells<br> | ||
| Line 391: | Line 474: | ||
- pMA_transmembraneregion_ß-Lactamase2<br> | - pMA_transmembraneregion_ß-Lactamase2<br> | ||
- pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker<br> | - pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker<br> | ||
| - | - pMA_Signalpeptide-Lipocalin_GGGS_N-CFP<br> | + | - pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F)<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_N-YFP<br> | + | - pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D)<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP<br> | + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E)<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP<br> | + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C)<br> |
<br> | <br> | ||
<h3>Oct. 15th 2008</h3> <br> | <h3>Oct. 15th 2008</h3> <br> | ||
<br> | <br> | ||
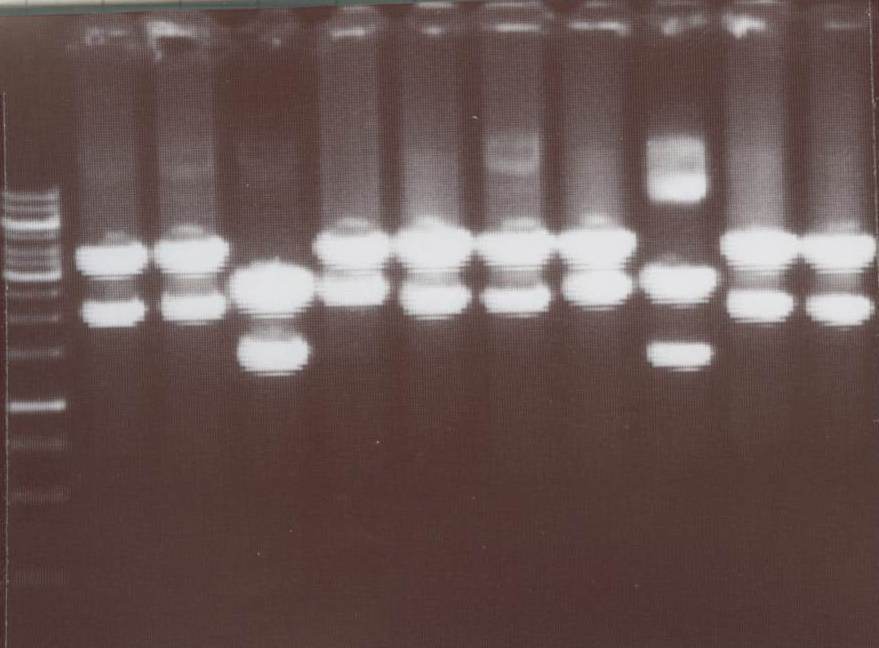
| - | '''Miniprep''' (Kathrin)<br> | + | '''Miniprep and analytic digestion''' (Kathrin)<br> |
| - | + | - pMA_transmembraneregion_ß-Lactamase1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase1 ~670bp <br> | |
| + | - pMA_transmembraneregion_ß-Lactamase2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase2 ~380bp<br> | ||
| + | - pMA_transmembraneregion_Luciferase 1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 1 ~ 350bp<br> | ||
| + | - pMA_transmembraneregion_Luciferase 2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 2 ~ 310bp<br> | ||
| + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) -> expected bands: pMA-Vector ~2360bp, Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) ~ 1150bp<br> | ||
| + | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC_1015.jpg|250px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: pMA_transmembraneregion_ß-Lactamase1 clone 1 (correct)<br> | ||
| + | Lane03: pMA_transmembraneregion_ß-Lactamase1 clone 2 (correct)<br> | ||
| + | Lane04: pMA_transmembraneregion_ß-Lactamase2 clone 1 (correct)<br> | ||
| + | Lane05: pMA_transmembraneregion_ß-Lactamase2 clone 2 (correct)<br> | ||
| + | Lane06: pMA_transmembraneregion_Luciferase 1 clone 1 (correct)<br> | ||
| + | Lane07: pMA_transmembraneregion_Luciferase 1 clone 2 (correct)<br> | ||
| + | Lane08: pMA_transmembraneregion_Luciferase 2 clone 1 (correct)<br> | ||
| + | Lane09: pMA_transmembraneregion_Luciferase 2 clone 2 (correct)<br> | ||
| + | Lane10: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 1 (correct)<br> | ||
| + | Lane11: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 2 (correct)<br> | ||
<br> | <br> | ||
| - | + | - pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) -> expected bands: pMA-Vector ~2360bp, Insert Lipo D ~ 1320bp<br> | |
| - | + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) -> expected bands: pMA-Vector ~2360bp, Insert Lipo E ~ 1140bp<br> | |
| + | - pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) -> expected bands: pMA-Vector ~2360bp, Insert Lipo F ~ 1320bp<br> | ||
| + | - pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker -> expected bands: pMA-Vector ~ 2360bp, Signalpeptide-scFv-anti-NIP_GGGS-Linker ~ 920bp<br> | ||
| + | - pMA-signalpeptide-scFv-anti-NIP -> expected bands: pMA-Vector ~ 2360bp, signalpeptide-Anti-Nip-sc-Fv ~ 880bp<br> | ||
| + | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC-F_1015.jpg|200px]]<br> | ||
| + | Lane01: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 1 (correct)<br> | ||
| + | Lane02: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 2 (correct) (correct)<br> | ||
| + | Lane03: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 1 (correct)<br> | ||
| + | Lane04: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 2 (correct)<br> | ||
| + | Lane05: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 1 (correct)<br> | ||
| + | Lane06: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 2 (correct)<br> | ||
| + | Lane07: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 1 (correct)<br> | ||
| + | Lane08: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 2 (correct)<br> | ||
| + | Lane09: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 3 (correct)<br> | ||
| + | Lane10: pMA-signalpeptide-scFv-anti-NIP (correct)<br> | ||
| + | Lane11: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
<br> | <br> | ||
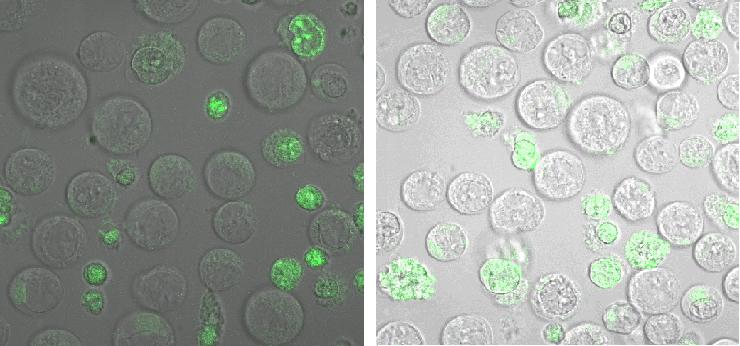
'''fluorescence microscopy''' (Normann and Kathrin) <br> | '''fluorescence microscopy''' (Normann and Kathrin) <br> | ||
| - | + | To test if the CMV-promotor in the transfectionvector allows a succesful transfection of a protein TV_CMV_5_YFP clone D was transfected to 293T cells and analysed by fluorescence microscopy. <br> | |
<br> | <br> | ||
[[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> | [[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> | ||
| + | '''Figure_fluorescence microscopy''' from left to right: 293T cells transfected with the Transfectionvektor+CMV-promotor and a YFP construct/ control/ brightfield picture of the control<br> | ||
| + | As expected the transfectionvector with CMV promotor induces expression of the yellow fluorescent protein.<br> | ||
<br> | <br> | ||
'''Preparative Digestion''' (Sabine)<br> | '''Preparative Digestion''' (Sabine)<br> | ||
| Line 434: | Line 550: | ||
of the ligations mentioned above<br> | of the ligations mentioned above<br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<h3>Oct. 16th 2008</h3> <br> | <h3>Oct. 16th 2008</h3> <br> | ||
| Line 484: | Line 562: | ||
<h3>Oct. 17th 2008</h3> <br> | <h3>Oct. 17th 2008</h3> <br> | ||
'''Miniprep''' (Sabine)<br> | '''Miniprep''' (Sabine)<br> | ||
| - | of NIP A,B and Lipo A,B, | + | of NIP A,B and Lipo A,B,G,H (both times clone 1 and 2)<br> |
| - | '''Analytic and Preparative Digestion''' | + | <br> |
| - | + | '''Analytic and Preparative Digestion''' (Sabine)<br> | |
| - | the analytic one with NotI, the preparative one with XbaI and PstI<br><br> | + | the analytic one with NotI, the preparative one with XbaI and PstI<br> |
| + | - Lipo A -> expected bands: pMA-Vector ~2360bp, Insert Lipo A ~ 1380bp<br> | ||
| + | - Lipo B -> expected bands: pMA-Vector ~2360bp, Insert Lipo B ~ 1090bp<br> | ||
| + | - Lipo G -> expected bands: pMA-Vector ~2360bp, Insert Lipo G ~ 1020bp<br> | ||
| + | - Lipo H -> expected bands: pMA-Vector ~2360bp, Insert Lipo H ~ 1060bp<br> | ||
| + | - NIP A -> expected bands: pMA-Vector ~2360bp, Insert NIP A ~ 1590bp<br> | ||
| + | - NIP B -> expected bands: pMA-Vector ~2360bp, Insert NIP B ~ 1300bp<br> | ||
| + | [[Image:TeamFreiburg2008_LipoA+B+G+H_NIPA+B_1017.jpg|150px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: Lipo A clone 1(correct)<br> | ||
| + | Lane03: Lipo B clone 1 (correct)<br> | ||
| + | Lane04: Lipo G clone 1 (correct)<br> | ||
| + | Lane05: Lipo H clone 1 (correct)<br> | ||
| + | Lane06: NIP A clone 1 (correct)<br> | ||
| + | Lane07: NIP B clone 1 (correct)<br> | ||
| + | <br> | ||
'''Miniprep''' (Michael)<br> | '''Miniprep''' (Michael)<br> | ||
of NIP C,D,E,F,G and H (each with clone 1 and 2)<br><br> | of NIP C,D,E,F,G and H (each with clone 1 and 2)<br><br> | ||
'''Analytic Digestion''' (Michael)<br> | '''Analytic Digestion''' (Michael)<br> | ||
| - | + | with NotI<br> | |
| + | - NIP C -> expected bands: pMA-Vector ~2360bp, Insert NIP C ~ 1360bp<br> | ||
| + | - NIP D -> expected bands: pMA-Vector ~2360bp, Insert NIP D ~ 1530bp<br> | ||
| + | - NIP E -> expected bands: pMA-Vector ~2360bp, Insert NIP E ~ 1360bp<br> | ||
| + | - NIP F -> expected bands: pMA-Vector ~2360bp, Insert NIP F ~ 1530bp<br> | ||
| + | - NIP G -> expected bands: pMA-Vector ~2360bp, Insert NIP G ~ 1230bp<br> | ||
| + | - NIP H -> expected bands: pMA-Vector ~2360bp, Insert NIP H ~ 1280bp<br> | ||
| + | [[Image:TeamFreiburg2008_NIPC-H_1017.jpg|200px]]<br> | ||
| + | Lane01: NIP C clone 1 (correct)<br> | ||
| + | Lane02: NIP D clone 1 (correct)<br> | ||
| + | Lane03: NIP E clone 1 (correct)<br> | ||
| + | Lane04: NIP F clone 1 (correct)<br> | ||
| + | Lane05: NIP G clone 1 (correct)<br> | ||
| + | Lane06: NIP H clone 1 (correct)<br> | ||
| + | Lane07: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | <br> | ||
'''Preparative Digestion''' (Michael)<br> | '''Preparative Digestion''' (Michael)<br> | ||
| - | of NIP C,D,E,F,G,H | + | of NIP C,D,E,F,G,H with XbaI and PstI<br><br> |
'''Picking clones''' (Sabine)<br> | '''Picking clones''' (Sabine)<br> | ||
Lipo III, IV, V and VI<br><br> | Lipo III, IV, V and VI<br><br> | ||
| Line 505: | Line 613: | ||
<h3>Oct. 18th 2008</h3> <br> | <h3>Oct. 18th 2008</h3> <br> | ||
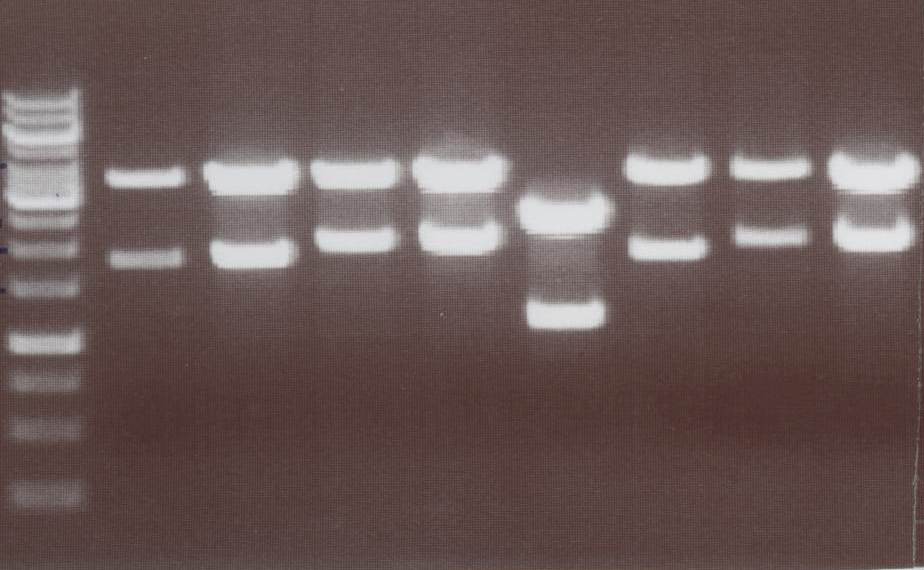
'''Miniprep and analytic digestion''' (Sabine)<br> | '''Miniprep and analytic digestion''' (Sabine)<br> | ||
| - | Lipo III, IV, V | + | - Lipo III -> expected bands: transfectionvector ~4100bp, Insert Lipo III (=Lipo C+CMV) ~ 1800bp<br> |
| + | - Lipo IV -> expected bands: transfectionvector ~4100bp, Insert Lipo IV (=Lipo D+CMV) ~ 1970bp<br> | ||
| + | - Lipo V -> expected bands: transfectionvector ~4100bp, Insert Lipo V (=Lipo E+CMV) ~ 1800bp<br> | ||
| + | - Lipo VI -> expected bands: transfectionvector ~4100bp, Insert Lipo VI (=Lipo F+CMV) ~ 1970bp<br> | ||
| + | [[Image:TeamFreiburg2008_Lipo3-4-5-6_1018.jpg|200px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: Lipo III clone 1 (correct)<br> | ||
| + | Lane03: Lipo III clone 2 (correct)<br> | ||
| + | Lane04: Lipo IV clone 1 (correct)<br> | ||
| + | Lane05: Lipo IV clone 2 (correct)<br> | ||
| + | Lane06: Lipo V clone 1 (not correct)<br> | ||
| + | Lane07: Lipo V clone 2 (correct)<br> | ||
| + | Lane08: Lipo VI clone 1 (correct))<br> | ||
| + | Lane09: Lipo VI clone 2 (correct)<br> | ||
<br> | <br> | ||
'''Ligation''' (Sabine) <br> | '''Ligation''' (Sabine) <br> | ||
| Line 534: | Line 655: | ||
<h3>Oct. 19th 2008</h3> <br> | <h3>Oct. 19th 2008</h3> <br> | ||
'''Miniprep and analytic digestion''' (Sabine)<br> | '''Miniprep and analytic digestion''' (Sabine)<br> | ||
| - | Lipo I, II, VII, VIII | + | - Lipo I -> expected bands: transfectionvector ~4100bp, Insert Lipo I (=Lipo A+CMV) ~ 2030bp<br> |
| + | - Lipo II -> expected bands: transfectionvector ~4100bp, Insert Lipo II (=Lipo B+CMV) ~ 1730bp<br> | ||
| + | - Lipo VII -> expected bands: transfectionvector ~4100bp, Insert Lipo VII (=Lipo G+CMV) ~ 1670bp<br> | ||
| + | - Lipo VIII -> expected bands: transfectionvector ~4100bp, Insert Lipo VIII (=Lipo H+CMV) ~ 1710bp<br> | ||
| + | [[Image:TeamFreiburg2008_Lipo1-2-7-8_1018.jpg|200px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: Lipo I clone 1 (correct)<br> | ||
| + | Lane03: Lipo I clone 2 (correct)<br> | ||
| + | Lane04: Lipo II clone 1 (correct)<br> | ||
| + | Lane05: Lipo II clone 2 (correct)<br> | ||
| + | Lane06: Lipo VII clone 1 (not correct)<br> | ||
| + | Lane07: Lipo VII clone 2 (correct)<br> | ||
| + | Lane08: Lipo VIII clone 1 (correct))<br> | ||
| + | Lane09: Lipo VIII clone 2 (correct)<br> | ||
| + | <br> | ||
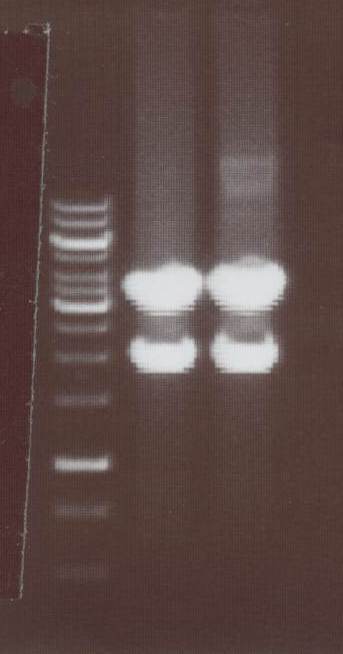
| + | - NIP I -> expected bands: transfectionvector ~4100bp, Insert NIP I (=NIP A+CMV) ~ 2240bp<br> | ||
| + | - NIP II -> expected bands: transfectionvector ~4100bp, Insert NIP II (=NIP B+CMV) ~ 1950bp<br> | ||
| + | [[Image:TeamFreiburg2008_Lipo1-2_1018.jpg|100px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: NIP I clone 1 (not correct)<br> | ||
| + | Lane03: NIP I clone 2 (correct)<br> | ||
| + | Lane04: NIP II clone 1 (correct)<br> | ||
| + | Lane05: NIP II clone 2 (correct)<br> | ||
<br> | <br> | ||
'''Picking clones''' <br> | '''Picking clones''' <br> | ||
| Line 543: | Line 686: | ||
-TV/CMV-CFP<br> | -TV/CMV-CFP<br> | ||
-TV/CMV-YFP<br> | -TV/CMV-YFP<br> | ||
| - | |||
-pMA-transmembrane-YFP<br> | -pMA-transmembrane-YFP<br> | ||
| + | -NIP III -> expected bands: transfectionvector ~4100bp, Insert NIP III (=NIP C+CMV) ~ 2010bp<br> | ||
| + | -NIP IV -> expected bands: transfectionvector ~4100bp, Insert NIP IV (=NIP D+CMV) ~ 2180bp<br> | ||
| + | -NIP V -> expected bands: transfectionvector ~4100bp, Insert NIP V (=NIP E+CMV) ~ 2010bp<br> | ||
| + | -NIP VI -> expected bands: transfectionvector ~4100bp, Insert NIP VI (=NIP F+CMV) ~ 2180bp<br> | ||
| + | -NIP VII -> expected bands: transfectionvector ~4100bp, Insert NIP VII (=NIP G+CMV) ~ 1880bp<br> | ||
| + | [[Image:TeamFreiburg2008_NIPIII-VIII_1018.jpg|200px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: NIP III clone 1 (correct)<br> | ||
| + | Lane03: NIP III clone 2 (correct)<br> | ||
| + | Lane04: NIP IV clone 1 (not correct)<br> | ||
| + | Lane05: NIP IV clone 2 (correct)<br> | ||
| + | Lane06: NIP VI clone 1 (correct)<br> | ||
| + | Lane07: NIP VI clone 2 (not correct)<br> | ||
| + | Lane08: NIP VII clone 1 (correct))<br> | ||
| + | Lane09: NIP VII clone 2 (correct)<br> | ||
| + | <br> | ||
| + | -NIP VIII -> expected bands: transfectionvector ~4100bp, Insert NIP VIII (=NIP H+CMV) ~ 1920bp<br> | ||
| + | [[Image:TeamFreiburg2008_NIPVIII_1020.jpg|70px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: NIP VIII clone 1 (correct)<br> | ||
| + | Lane03: NIP VIII clone 2 (correct)<br> | ||
<br> | <br> | ||
<h3>Oct. 21th 2008</h3> <br> | <h3>Oct. 21th 2008</h3> <br> | ||
| Line 572: | Line 735: | ||
-YFP, CFP, transmembrane-YFP clone 1 and 2, pMA-transmembrane, NIP I and Lipo I<br> | -YFP, CFP, transmembrane-YFP clone 1 and 2, pMA-transmembrane, NIP I and Lipo I<br> | ||
<br> | <br> | ||
| + | '''Gelextraction of the digested constructs'''(Normann)<br>using QIAquick gel extraction kit<br><br> | ||
| + | |||
'''Miniprep''' (Kathrin)<br> | '''Miniprep''' (Kathrin)<br> | ||
-of the 5ml cultures of the sequenced parts<br> | -of the 5ml cultures of the sequenced parts<br> | ||
-of the 20ml cultures of the parts needed for transfection<br> | -of the 20ml cultures of the parts needed for transfection<br> | ||
<br> | <br> | ||
| - | '''Phage DNA Isolation''' (Normann)<br> | + | '''Phage DNA Isolation''' (Normann)<br>Dilution of the 50ml culture to an OD of approximately 0,1<br> |
| - | Dilution of the 50ml culture to an OD of approximately 0,1<br> | + | <br>'''Doubletransfection of 293t-cells'''(Normann) |
| + | <br>done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system and the two parts of the anti NIP splitluc- receptor system. | ||
| + | |||
<br> | <br> | ||
'''Ligation''' (Kathrin)<br> | '''Ligation''' (Kathrin)<br> | ||
| Line 588: | Line 755: | ||
Inoculate ER2738 cells with M13 phages -> shaking for approximately 4h at 37°C<br> | Inoculate ER2738 cells with M13 phages -> shaking for approximately 4h at 37°C<br> | ||
<br> | <br> | ||
| + | After that, spinning down 50 ml at 5000g 20min and adding 7ml PEG/NaCl and storing at 4°C. Incubation over night. | ||
<h3>Oct. 23th 2008</h3> <br> | <h3>Oct. 23th 2008</h3> <br> | ||
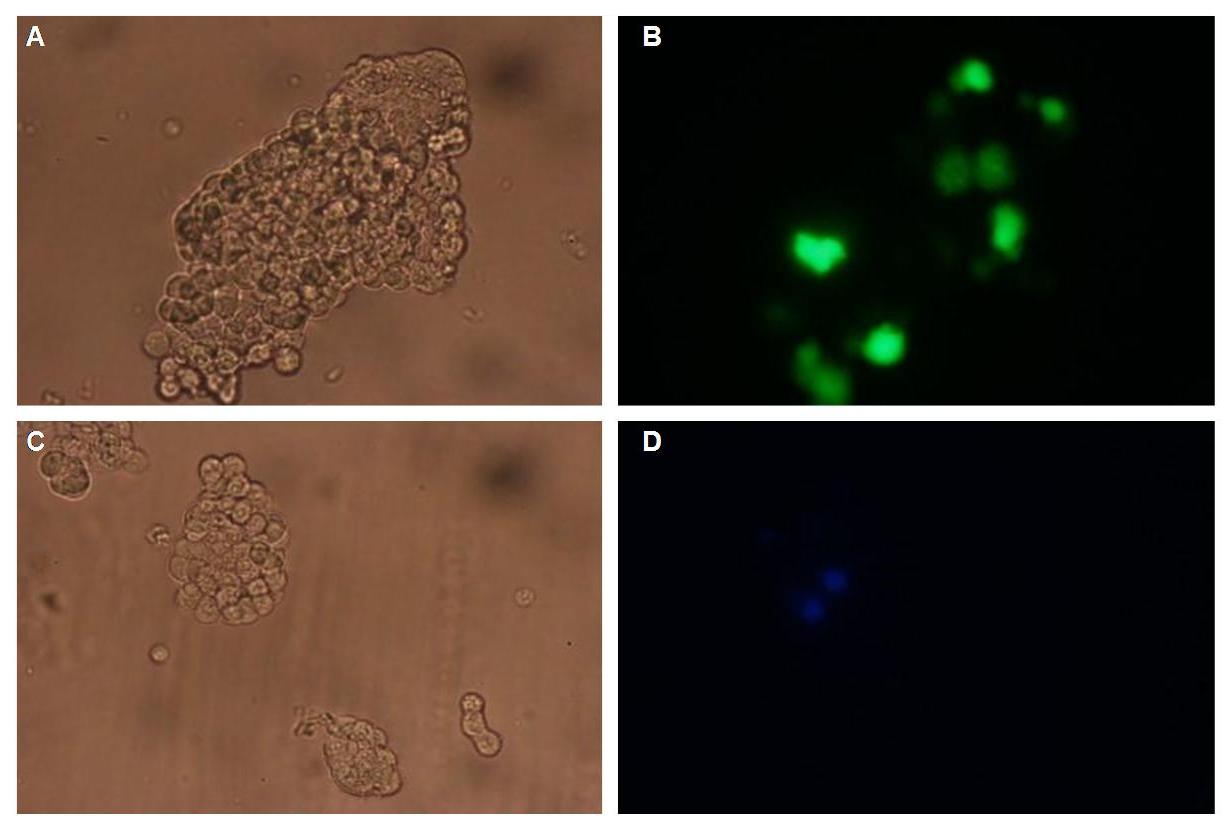
| + | '''Detecting the new origami at the AFM (Normann)''' <br> | ||
| + | [[Image:Origamis2FreiGEM08.JPG]]<br> | ||
'''Sequencing''' (Kathrin)<br> | '''Sequencing''' (Kathrin)<br> | ||
Correct:<br> | Correct:<br> | ||
| Line 594: | Line 764: | ||
-NIP C, D and H clones 2<br> | -NIP C, D and H clones 2<br> | ||
<br> | <br> | ||
| + | >'''Doubletransfection of 293t-cells'''(Normann) | ||
| + | <br>done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system, the two parts of the anti NIP splitluc- receptor system and the two parts of the anti NIP split GFP receptor system.<br><br> | ||
'''Preparative Digestion''' (Kathrin)<br> | '''Preparative Digestion''' (Kathrin)<br> | ||
NIP C, D and H (clones 2)<br> | NIP C, D and H (clones 2)<br> | ||
| Line 629: | Line 801: | ||
<h3>Oct. 24th 2008</h3> <br> | <h3>Oct. 24th 2008</h3> <br> | ||
'''Analytic digestion''' (Sabine)<br> | '''Analytic digestion''' (Sabine)<br> | ||
| - | NIP I | + | - NIP I+YFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+YFP(=NIP A+CMV+YFP) ~ 2940bp<br> |
| + | - NIP I+CFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+CFP(=NIP A+CMV+CFP) ~ 2940bp<br> | ||
| + | - Lipo I+YFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+YFP (=Lipo A+CMV+YFP) ~ 2730bp<br> | ||
| + | - Lipo I+CFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+CFP (=Lipo A+CMV+CFP) ~ 2730bp<br> | ||
| + | [[Image:TeamFreiburg2008_NIP+YFPusw_1024.jpg|250px]]<br> | ||
| + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> | ||
| + | Lane02: NIP I+YFP clone 1 (correct)<br> | ||
| + | Lane03: NIP I+YFP clone 2 (correct)<br> | ||
| + | Lane04: NIP I+YFP clone 3 (correct)<br> | ||
| + | Lane05: NIP I+CFP clone 1 (correct)<br> | ||
| + | Lane06: NIP I+CFP clone 2 (correct)<br> | ||
| + | Lane07: ---<br> | ||
| + | Lane08: Lipo I+YFP clone 1 (correct)<br> | ||
| + | Lane09: Lipo I+YFP clone 2 (correct)<br> | ||
| + | Lane10: Lipo I+CFP clone 1 (correct)<br> | ||
| + | <br> | ||
| + | '''staining cells with LiveBLAzer from invitrogen'''(Normann,Michael) | ||
| + | <br> The cells which should have expressed both parts of the anti NIP bla receptor system were stained with LiveBLAzer, using the protocol of the invitrogen LiveBLAzer Kit.<br><br> | ||
'''Preparative digestion''' (Sabine)<br> | '''Preparative digestion''' (Sabine)<br> | ||
-pMA-Signalpeptide-scFV-Anti-NIP-transmembrane-YFP<br> | -pMA-Signalpeptide-scFV-Anti-NIP-transmembrane-YFP<br> | ||
| Line 694: | Line 883: | ||
<br> | <br> | ||
<h3>Oct. 27th 2008</h3> <br> | <h3>Oct. 27th 2008</h3> <br> | ||
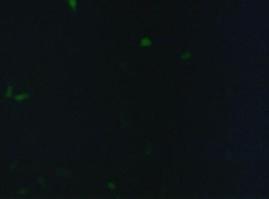
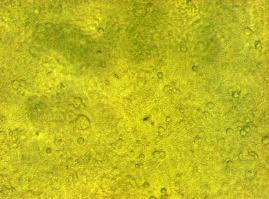
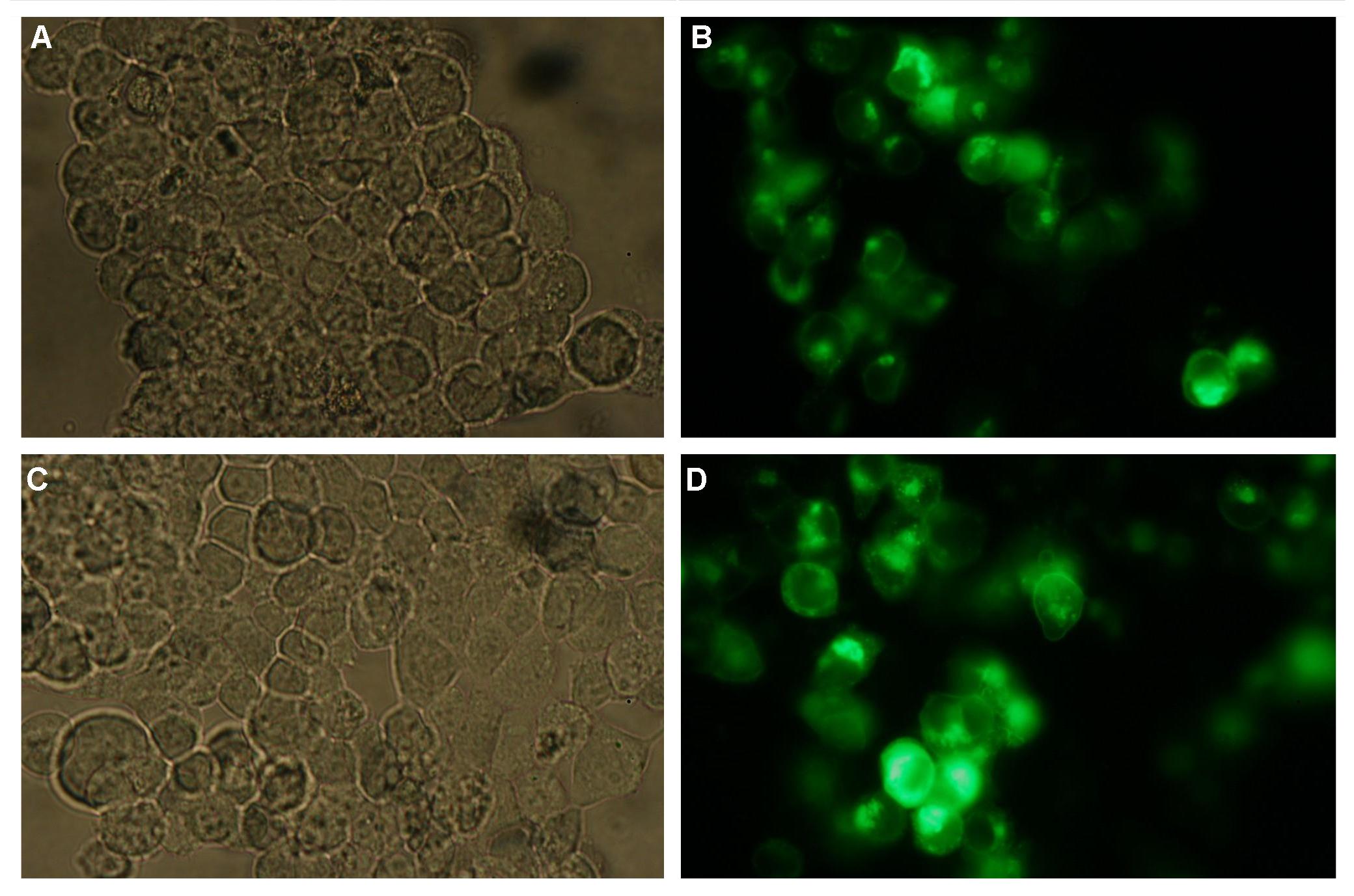
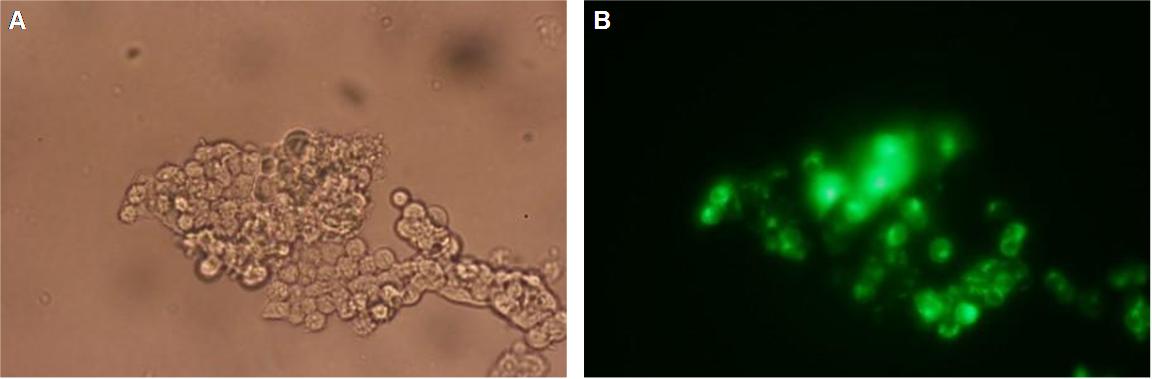
| + | '''fluorescence microscopy''' (Michael, Sabine, Kathrin)<br> | ||
| + | analysis of the transfected cells 1)-12) via fluorescence microscopy.<br> | ||
| + | 1) no result <br> | ||
| + | 2) no result <br> | ||
| + | 3) no result <br> | ||
| + | 4) no result <br> | ||
| + | 5) no result <br> | ||
| + | 6) no result <br> | ||
| + | 7) as expected <br> | ||
| + | 8) as expected <br> | ||
| + | 9) as expected <br> | ||
| + | 10) as expected <br> | ||
| + | 11) as expected <br> | ||
| + | 12) as expected <br><br> | ||
| + | |||
| + | Analysis of the split beta Lactamase was performed by LiveBLAzer (invitrogen). After loading the cells with CCF4-AM the molecule is converted into CCF4 by an esterase in the cell. Beta Lactamase splits the FRET-pair CCF4 (absoption 409, emission 520nm) into two fragments. One of the fragments can still be stimulated by 409nm to detect an emission at 447nm)<br><br> | ||
| + | |||
| + | [[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP 1.jpg|600px]]<br> | ||
| + | <br><br> | ||
| + | [[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP 2.jpg|600px]]<br> | ||
| + | '''Figure_fluorescence microscopy 1''' brightfield and UV-light picture with YFP-filter. The 293T cells are transfected with the construct: Transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP.<br><br><br> | ||
| + | [[Image:Freiburg2008_TV_CMV_YFP CFP_loeslich.jpg|600px]]<br> | ||
| + | '''Figure_fluorescence microscopy 2''' brightfield and UV-light picture with YFP- and CFP-filter. The 293T cells are transfected with the constructs: '''upper:'''Transfectionsvector+CMV_YFP '''lower:'''Transfectionsvector+CMV_CFP<br><br> | ||
| + | |||
| + | The pictures show cleary the difference between soluble YFP/CFP '''Figure_fluorescence microscopy 2''' and the membranelocalised YFP fused to the synthetic receptor construct '''Figure_fluorescence microscopy 1''' "transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP". The principle to localise our synthetic fusion-receptor-proteins to cell membrane is proved. <br> | ||
| + | |||
| + | <!--<h3>Oct. 28th 2008</h3> <br>--> | ||
| - | |||
}} | }} | ||
Latest revision as of 02:40, 30 October 2008
 "
"