Team:Heidelberg/Notebook/Killing II/10thweek
From 2008.igem.org
(Difference between revisions)
(→Wednesday 10/08/2008) |
(→Sunday 10/12/2008) |
||
| (3 intermediate revisions not shown) | |||
| Line 685: | Line 685: | ||
====Sender-activity-test==== | ====Sender-activity-test==== | ||
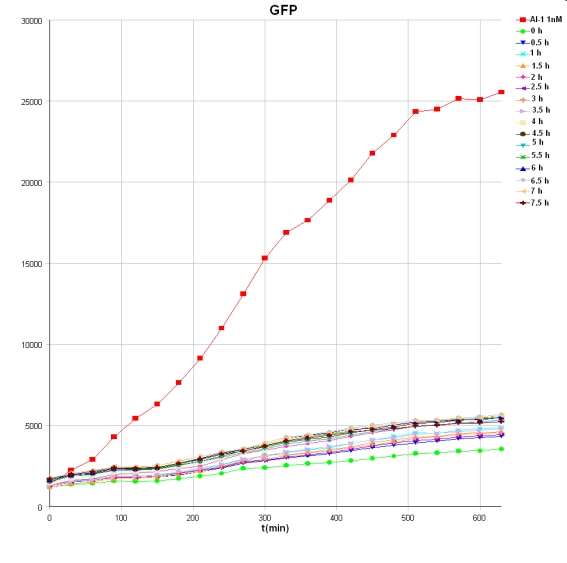
*Results of ON measurements: The graph shows the GFP intensities over time for the different supernatants and one AI-1 concentration (c = 1 nM). The GFP induction is much lower for the supernatants in comparison to the AI-1. Other tests with lower AI-1 concentration have to be performed to determine the AI-1 production rate. [[Image: 081008-sender_Activity_test_GFP_sup_besch_small.jpg | 700 px | center ]] | *Results of ON measurements: The graph shows the GFP intensities over time for the different supernatants and one AI-1 concentration (c = 1 nM). The GFP induction is much lower for the supernatants in comparison to the AI-1. Other tests with lower AI-1 concentration have to be performed to determine the AI-1 production rate. [[Image: 081008-sender_Activity_test_GFP_sup_besch_small.jpg | 700 px | center ]] | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/10thweek back]] | ||
==Friday 10/10/2008== | ==Friday 10/10/2008== | ||
| Line 709: | Line 711: | ||
===Sender Cloning: constitutive promotor - sender=== | ===Sender Cloning: constitutive promotor - sender=== | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/10thweek back]] | |
| - | + | ||
==Saturday 10/11/2008== | ==Saturday 10/11/2008== | ||
| Line 868: | Line 869: | ||
**plate scheme: <br> [[Image: 081011-plate_scheme_sender_amplifier_test.jpg | 800 px | center]] | **plate scheme: <br> [[Image: 081011-plate_scheme_sender_amplifier_test.jpg | 800 px | center]] | ||
**Unfortunately we were not able to measure ON because the Tecan plate reader was not recognized by the PC. | **Unfortunately we were not able to measure ON because the Tecan plate reader was not recognized by the PC. | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/10thweek back]] | ||
==Sunday 10/12/2008== | ==Sunday 10/12/2008== | ||
| Line 1,016: | Line 1,019: | ||
- Plate 500 µl on preheated LB-Amp plate | - Plate 500 µl on preheated LB-Amp plate | ||
- incubate overnight | - incubate overnight | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/10thweek back]] | ||
| + | |||
[[Team:Heidelberg/Notebook/Killing_II/11thweek | go to 11<sup>th</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/11thweek | go to 11<sup>th</sup> week]] | ||
Latest revision as of 21:44, 28 October 2008


10th week
Contents |
Monday 10/06/2008
pSB1A2-Receiver-Colicin cloning
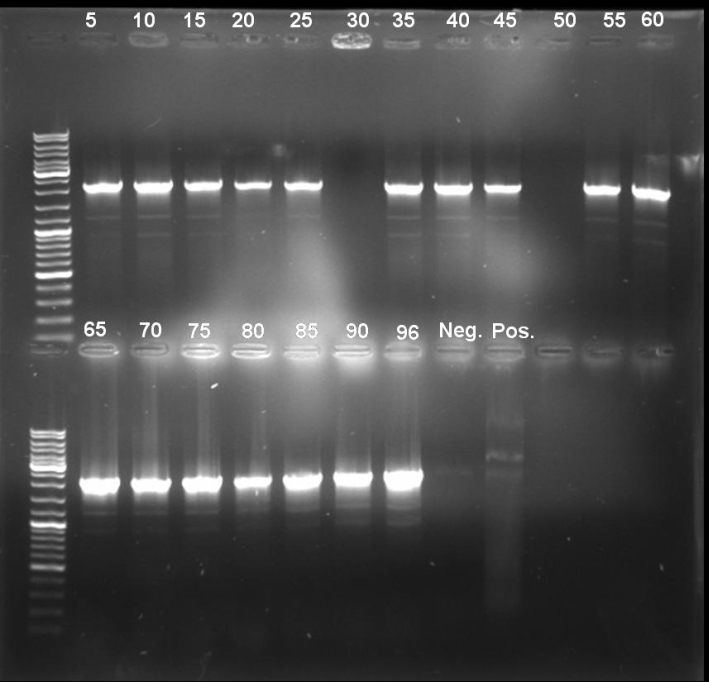
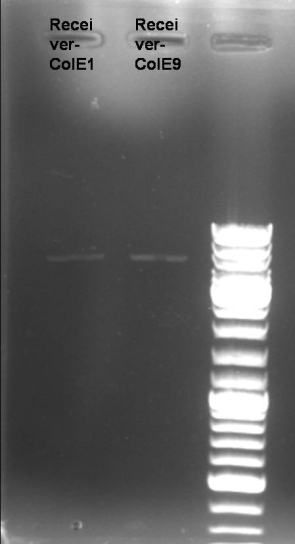
- Gel of E9 lys PCR-Screen: 1% Agarose, 135 V, 35 min
- Gel-Results of E9-PCR-Screen:
- Estimated Bands: ~2150bp
- Estimated Bands visible in many tubes -> Screening of single colonies.
- PCR-Screen of single colonies of pSB1A2-Receiver-Col E9 lys
25.0 µl Taq Master Mix (Fermentas) 2.5 µl T9002_LuxpR_Not_Eco_Xba_G_fw (Tm=80,96°C) 2.5 µl ColE9_lyProt_rv_A_SpeI (Tm=64,29°C) 20.0 µl H2O 1 colony ------- 50.0 µl
program:
95 °C 3 min 95 °C 1 min | 64 °C 1 min | 30 cycles 72 °C 1 min 30 sec | 72 °C 10 min 4 °C constant
- Gel of E9 lys PCR-Screen: 1% Agarose, 135 V, 35 min
- Gel-Results of E9-PCR-Screen: Colonies 1, 2, 3, 4, 7 & 8 positive; Inoculation of 5 ml LB-Amp media with this colonies for miniprep, 37 °C -> ON
Colicin Activity Test
- Plan for activity tests of Colicin-Receivers:
- Overnight measurement (Monday - Tuesday) in Tecan plate reader with reference promotor cells (BBa_I20260) in combination with pSB1A2-Rec-ColE9 lys lysed supernatant.
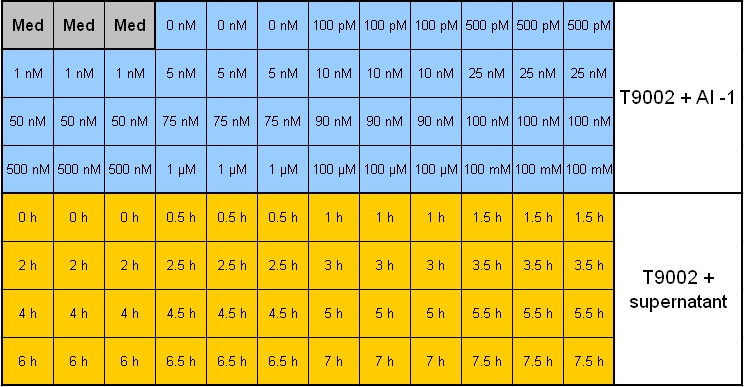
- Plate scheme (click for detailed image):
[back]
Tuesday 10/07/2008
pSB1A2-Receiver-Colicin cloning
- Colicin-Activity-Test: Cultures were not grown -> new inoculation
- Miniprep of pSB1A2-Receiver-ColE9lys (BioBrick Standard): colonies 1, 2, 3, 4, 7 & 8, eluted in 35 µl H2O
- Digestion of pSB1A2-Receiver-ColE9lys (BioBrick Standard): colonies 1, 2, 3, 4, 7 & 8
colonies 1 & 8:
5.0 µl DNA (~65-80 ng/µl) 25.5 µl H2O 4.0 µl BSA 10x (NEB) 4.0 µl NEBuffer 2 (NEB) 0.5 µl SpeI (NEB) 1.0 µl XbaI (NEB) ------- 40.0 µl
colonies 2, 3 & 4:
2.0 µl DNA (~245-275 ng/µl) 28.5 µl H2O 4.0 µl BSA 10x (NEB) 4.0 µl NEBuffer 2 (NEB) 0.5 µl SpeI (NEB) 1.0 µl XbaI (NEB) ------- 40.0 µl
colony 7:
2.0 µl DNA (~125 ng/µl) 28.5 µl H2O 4.0 µl BSA 10x (NEB) 4.0 µl NEBuffer 2 (NEB) 0.5 µl SpeI (NEB) 1.0 µl XbaI (NEB) ------- 40.0 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Digestion results:
- Expected fragments:
- 2 x ~2000 (insert & vector), if XbaI site is recovered
- 1 x ~4000, if XbaI site is not recovered
- In each colony there is only one fragment. Probably we have no positive clones with the BioBrick standard. To check the sequence we send the probes for sequencing.
- Expected fragments:
- Send probes 1, 2, 3, 4, 7, & 8 to GATC for sequencing: Primers VF2 & VR
- Growth test of each Rec-Colicin glycerol stock: Inoculation in 2 ml LB-Amp and 2 ml TB-Amp. 37 °C -> ON
Sender cloning: constitutive promotor-sender
Sender-activity-test
- Inoculation of 15 ml TB-Amp media with sender (J23107 + F1610) from glycerol stock
- Inoculation of 15 ml TB-Amp media with GFP-receiver ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002])
- Inoculation of 15 ml LB-Amp media with sender (J23107 + F1610) from glycerol stock
- Inoculation of 15 ml LB-Amp media with GFP-receiver ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002])
- Plan for Wednesdaynesday:
- Inoculation of 7 ml TB media with 150 µl from sender ONC
- Inoculation of 7 ml TB media with 7 ml from GFP-receiver ONC
- Count amount of cells from ONC dilution t = 0 h (counting chamber)
- Measurement of optical density (OD) of ONC dilution t = 0 h
- Measurement of OD of one probe half-hourly for 8 - 10 h
- Creation of supernatant the measured probe by centrifugation and filtration half-hourly for 8 - 10 h
- Storing supernatant at 4 °C
- Count amount of cells from last probe (counting chamber)
- Start overnight measurement in Tecan plate reader
- plate scheme (click for detailed image):
[back]
Wednesday 10/08/2008
pSB1A2-Receiver-Colicin cloning
Standardization of pSB1A2-colE9lys-Receiver
- Sequencing results: Colonies 1, 2, 3, 4 & 8 have the right sequence. Our first colicin Receiver fullfills BioBrick standard and is ready for activity tests. Colony 7 has mistakes inside the receiver and is thrown away.
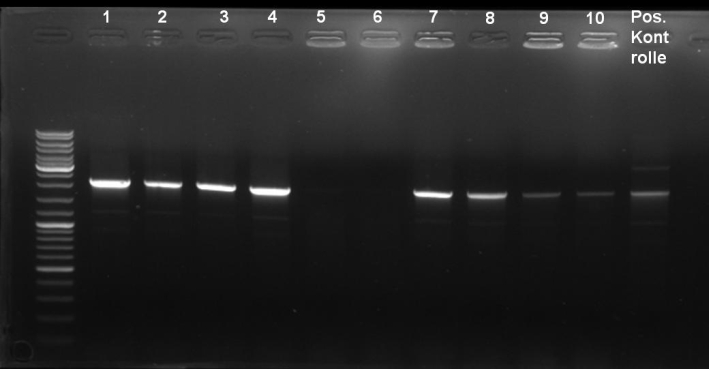
- Digestion of pSB1A2-Receiver-ColE9lys colony 2 with XbaI/SpeI, XbaI, SpeI, EcoRI/PstI:
'XbaI/SpeI:' 5.0 µl DNA (~240 ng/µl) 5.0 µl NEBuffer 2 (NEB) 5.0 µl BSA 10x (NEB) 0.5 µl XbaI (20 000 Units/ml, NEB) 1.0 µl SpeI (10 000 Units/ml, NEB) 33.5 µl H2O ------- 50.0 µl 'XbaI:' 5.0 µl DNA (~240 ng/µl) 5.0 µl NEBuffer 2 (NEB) 5.0 µl BSA 10x (NEB) 0.5 µl XbaI (20 000 Units/ml, NEB) 34.5 µl H2O ------- 50.0 µl
'SpeI:' 5.0 µl DNA (~240 ng/µl) 5.0 µl NEBuffer 2 (NEB) 5.0 µl BSA 10x (NEB) 1.0 µl SpeI (10 000 Units/ml, NEB) 34.0 µl H2O ------- 50.0 µl
'EcoRI/PstI:' 5.0 µl DNA (~240 ng/µl) 5.0 µl NEBuffer 2 (NEB) 5.0 µl BSA 10x (NEB) 0.5 µl EcoRI (20 000 Units/ml, NEB) 0.5 µl PstI (20 000 Units/ml, NEB) 34.0 µl H2O ------- 50.0 µl
Colicin Acitivity test
- Inoculation of ONC for colicin activity test on Thursday, 2008-10-09:
- 10 ml TB-Kana with reference promoter cells from glycerolstocks ([http://partsregistry.org/Part:BBa_I20260 BBa_I20260])
- 10 ml TB-Amp with pSB1A2-Receiver, pSB1A2-Receiver-ColE9lys, pSB1A2-Receiver-ColE1 from Glycerolstocks
Sender cloning: constitutive promotor-sender
Sender-activity-test
- Inoculation of 7 ml TB media with 150 µl from sender ONC
- Inoculation of 7 ml TB media with 7 ml from GFP-receiver ONC
- Measurement of optical density (OD) of ONC dilution t = 0 h
- Measurement of OD of one probe half-hourly for 7 h
- Creation of supernatant the measured probe by centrifugation and filtration half-hourly for 8 - 10 h
- Storing supernatant at 4 °C
- Start overnight measurement in Tecan plate reader
- plate scheme (click for detailed image): [[Image: 081007-pipetting_scheme_sender_test.jpg | 800 px | center ]
[back]
Thursday 10/09/2008
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-Receiver-Colicin E1/E9
- Mutagenesis PCR:
5.0 µl pfu buffer 1.0 µl forward primer (10 µM) 1.0 µl reverse primer (10 µM) 1.0 µl dNTPs (12.5 mM, 50x) 0.5 µl template DNA - 5 to 50 ng plasmid DNA 40.5 µl milliQ water 1.0 µl cloned pfu polymerase (Stratagene)
95 °C 30 sec 95 °C 30 sec | 55 °C 60 sec | 16 cycles 68 °C 11 min | 4 °C constant
- Digestion with DpnI (NEB) to cut parental plasmid: Added 1 µl of DpnI to the finished Mutagenesis PCR. DpnI cuts at methylated GATC sites, so that only parental plasmids without the mutation are cutted. 2 h -> 37 °C
- PCR Purification Kit (Qiagen) to purify the plasmids.
- Transformation of prior mutagenesis: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 200 µl on preheated LB-Amp plate - incubate overnight
- Controlgel of Mutagenesis: 1 % Agarose, 135 V, 30 min; DNA is visible -> probably the mutagenesis worked.
Colicin Activity tests
- 10 am: Inocullation of TB media from ONC
- 50 ml TB-Kana with 1.4 ml ONC of reference promoter cells ([http://partsregistry.org/Part:BBa_I20260 BBa_I20260])
- 3 x 50 ml TB-Amp with 1.4 ml ONC of pSB1A2-Receiver, pSB1A2-Receiver-ColE9lys, pSB1A2-Receiver-ColE1
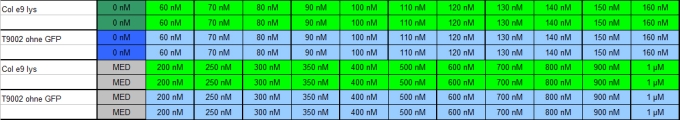
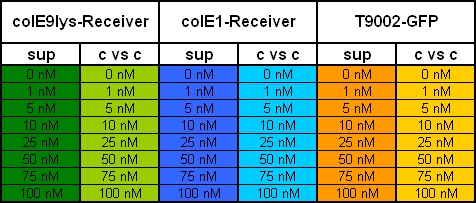
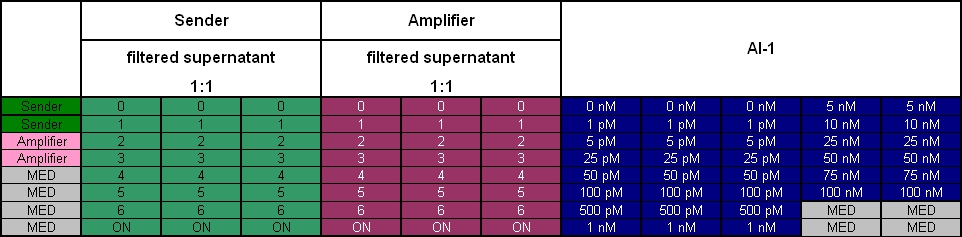
- 12.30 am: OD -> 0.320. Separation of the 3 Receiver cultures into 8 tubes with 3 ml. Activation with 8 different Autoinducer([http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/K3007 detailed information, Sigma]) concentrations: 0 nM, 1 nM, 5 nM, 10 nM, 25 nM, 50 nM, 75 nM & 100 nM.
- 8.00 pm: Preparing the test
- plate scheme:
- Preparation of probes:
- 10 pm start of measurement
- plate scheme:
Sender cloning: constitutive promotor-sender
Sender-activity-test
- Results of ON measurements: The graph shows the GFP intensities over time for the different supernatants and one AI-1 concentration (c = 1 nM). The GFP induction is much lower for the supernatants in comparison to the AI-1. Other tests with lower AI-1 concentration have to be performed to determine the AI-1 production rate.
[back]
Friday 10/10/2008
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-Receiver-Colicin E1/E9
- Inoculation of liquid cultures from transformation of the first mutagenesis.
- Miniprep of liquidcultures: eluted in 35 µl H2O (Qiagen Miniprepkit)
- 2nd mutagenesis PCR of Colicin E1 - Receiver (mutation of first PstI site):
- Mutagenesis PCR:
5.0 µl pfu buffer 1.0 µl forward primer (10 µM) 1.0 µl reverse primer (10 µM) 1.0 µl dNTPs (12.5 mM, 50x) 0.5 µl template DNA - 5 to 50 ng plasmid DNA 40.5 µl milliQ water 1.0 µl Turbo pfu polymerase (Stratagene)
95 °C 30 sec 95 °C 30 sec | 55 °C 60 sec | 16 cycles 68 °C 11 min | 4 °C constant
Sender Cloning: constitutive promotor - sender
[back]
Saturday 10/11/2008
Construction of reporterplasmid with kanamycin resistance
mCherry
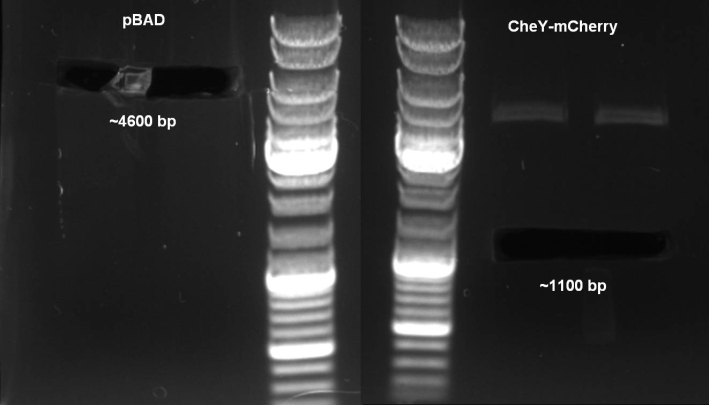
- For the visualization of our killer cells we need a RFP or mCherry reporter plasmid with kanamycin resistance. We decided to clone a cheY-mCherry fusion protein into pBAD18 vector.
- Digestion of pBAD18 plasmid & pES16-CheY-mCherry with SacI/XbaI: 2h 37 °C, 20 min 65 °C
20.0 µl DNA 0.5 µl SacI (20 000 U/ml, 100 % Activity in NEBuffer 4, NEB) 0.7 µl XbaI (20 000 U/ml, 75 % Activity in NEBuffer 4, NEB) 5.0 µl BSA 10x 5.0 µl NEBuffer 4 (NEB) 18.8 µl H2O ------- 50.0 µl
- Gel of Digestion: 0.7% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments:
- pBAD -> ~4600 bp
- CheY-mCherry -> ~1100 bp
- We were able to cut both fragments.
- Gelextraction with Qiagen Kit: eluted in 35 µl H2O
- Expected Fragments:
- Ligation of pBAD & mCherry: 11 h -> 16 °C, 20 min -> 65 °C
upper band
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 0.5 µl pBAD (11.4 ng/µl) 15.5 µl CheY-mCherry (8.3 ng/µl) ------- 20.0 µl
- EDIT 10/12/202008: calculated DNA ratio wrong
RFP
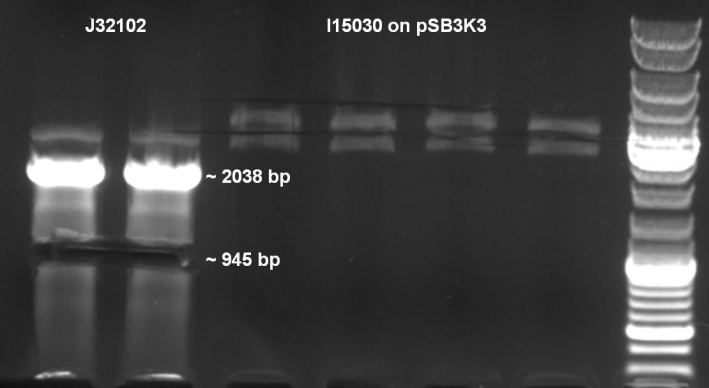
- In addition we try to clone RFP gene cassette from J23102 into pSB3K3.
- Digestion of BBa_J23102 & BBa_I15030 with EcoRI/PstI: 2 h -> 37 °C, 20 min -> 65 °C
20.0 µl DNA 1.0 µl EcoRI (20 000 U/ml, NEB) 1.0 µl PstI (10 000 U/ml, NEB) 5.0 µl BSA 10x 5.0 µl EcoRI Buffer(NEB) 18.5 µl H2O ------- 50.0 µl
- Gel of Digestion: 0.7% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments:
- J23102 -> ~945 bp
- I15030 -> ~1909 bp
- pSB3K3 -> ~2750 bp
- J23102 was cutted out of the gel. For pSB3K3 we were not sure which was the right fragment so we go on with both fragments.
- Gelextraction with Qiagen Kit: eluted in 35 µl H2O
- Expected Fragments:
- Ligation of J23102 and pSB3K3 (5:1):
upper band
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 8.5 µl pSB3K3 (upper Band) 7.5 µl BBa_J23102 ------- 20.0 µl
lower band
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 11.6 µl pSB3K3 (lower band) 4.4 µl BBa_J23102 ------- 20.0 µl
- EDIT 10/12/202008: calculated DNA ratio wrong
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-Receiver-Colicin E1/E9
- Digestion of overnight PCR probes with DpnI for 2 h -> 37 °C.
- PCR purification of probes: eluted in 35 µl H2O (Qiagen Kit)
- Transformation of prior mutagenesis: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 200 µl on preheated LB-Amp plate - incubate overnight
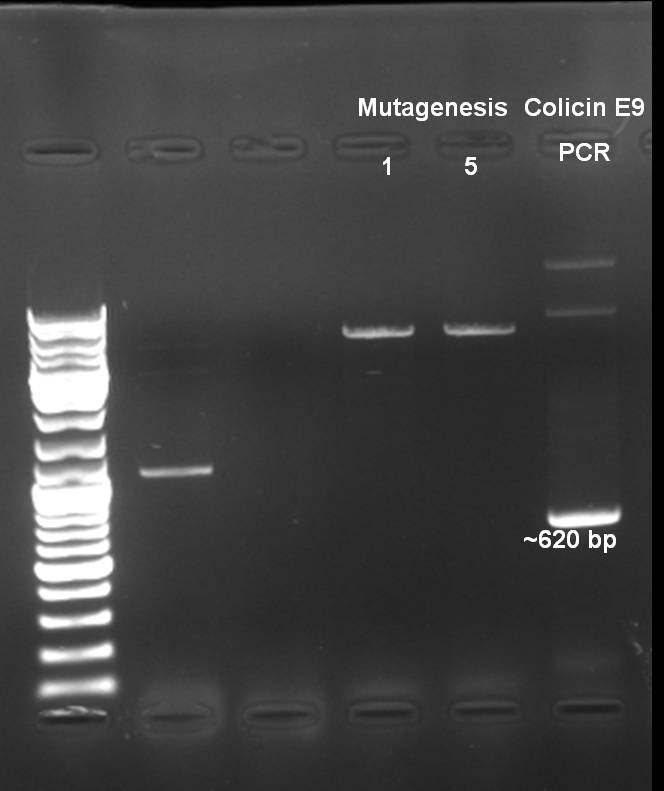
- Controlgel of Mutagenesis: 1 % Agarose, 135 V, 30 min; DNA is visible -> probably the mutagenesis worked.
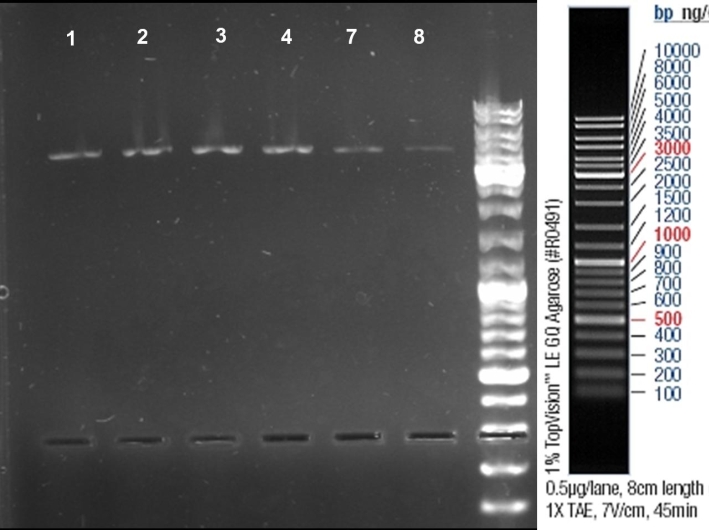
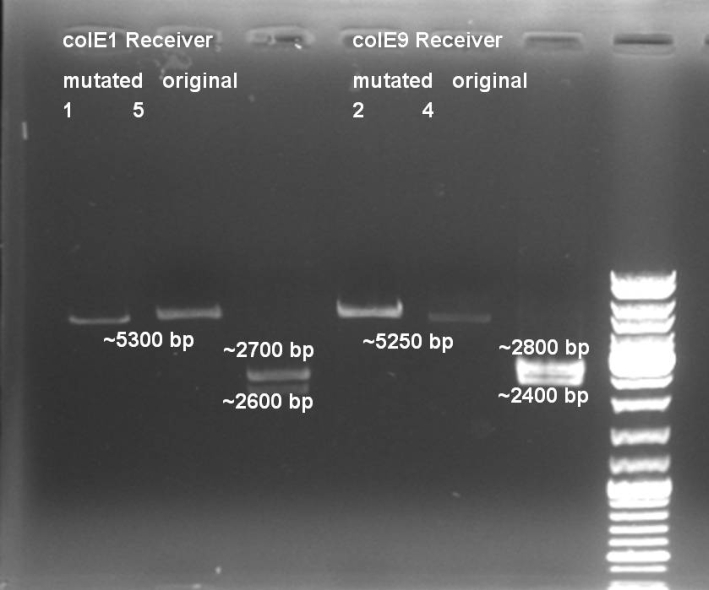
- Controlldigestion of first mutagenesis of Colicin E1 & E9 - Receiver with EcoRI: 2h -> 37 °C
10.0 µl DNA 2.0 µl BSA 10x (NEB) 0.5 µl EcoRI (NEB) 2.0 µl Buffer EcoRI 5.5 µl H2O ------- 20.0 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Gelresults: Mutated Colicin E1 and E9 Receivers were cutted only once by EcoRI. -> Mutagenesis sucessful.
Colicin E9 operon - parts: Colicin E9
- Digestion of Colicin E9 PCR product & pSB3K3 (from BBa_I15030)
20.0 µl DNA 1.0 µl EcoRI (20 000 U/ml, NEB) 1.0 µl PstI (10 000 U/ml, NEB) 5.0 µl BSA 10x 5.0 µl EcoRI Buffer(NEB) 18.5 µl H2O ------- 50.0 µl
- Gel of Digestion: 0.7% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments:
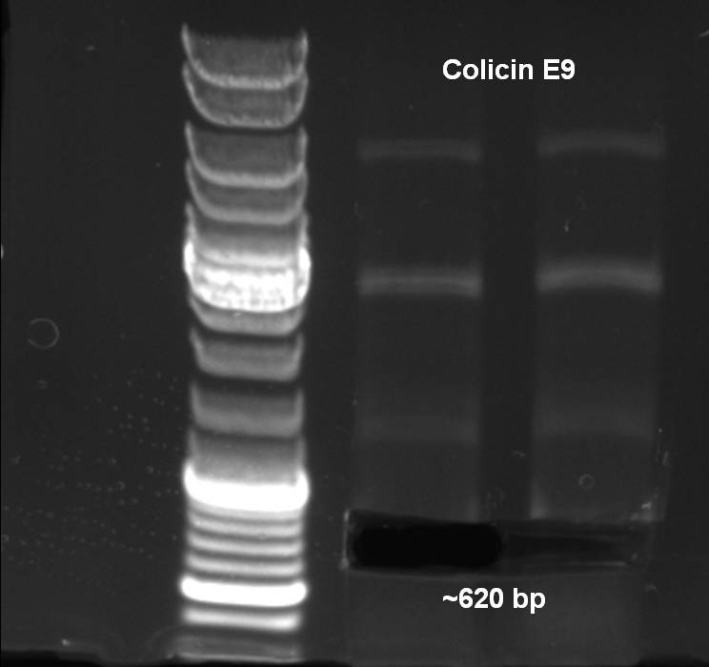
- Colicin E9 -> ~620 bp
- Colicin E9 was cutted out of the gel.
- Gelextraction with Qiagen Kit: eluted in 35 µl H2O
- Expected Fragments:
- Ligation of colicin E9 and pSB3K3 (5:1):
upper band
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 6.1 µl pSB3K3 (upper Band) 9.9 µl Colicin E9 ------- 20.0 µl
lower band
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 9.5 µl pSB3K3 (lower band) 8.5 µl Colicin E9 ------- 20.0 µl
EDIT 10/12/202008: calculated DNA ratio wrong
Sender Cloning: constitutive promotor - sender
Sender activity test
- Constitutive sender and amplifier ([http://partsregistry.org/Part:BBa_I15030 BBa_I15030]) activity test
- 7x Inoculation of 7 ml TB media with 140 µl from sender ONC
- 7x Inoculation of 7 ml TB media with 140 µl from amplifier ONC
- Inoculation of 7 ml TB media with 7 ml from GFP-receiver ONC
- Measurement of optical density (OD) of ONC dilutions t = 0 h
- Measurement of OD of one probe hourly for 6 h
- Creation of supernatant of the measured probe by centrifugation and filtration hourly for 6 h
- Storing supernatant at 4 °C
- Count amount of cells from last probe (counting chamber)
- plate scheme:
- Unfortunately we were not able to measure ON because the Tecan plate reader was not recognized by the PC.
[back]
Sunday 10/12/2008
Construction of reporterplasmid with kanamycin resistance
mCherry
- Transformation of pBAD-mCherry Ligation of saturday, 10/11/2008 (wrong vector-insert ratio): 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight at 37°C
- Ligation of pBAD and mCherry (1:5): 1 h -> 22 °C, 20 min -> 65 °C
- Reason: Transformation on Saturdayurday was carried out with wrong ratio of vector and insert (calculation error).
2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 6.05 µl pBAD (11.4 ng/µl) 9.95 µl CheY-mCherry (8.3 ng/µl) ------- 20.0 µl
- Transformation of pBAD-mCherry Ligation: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight at 37°C
RFP
- Transformation of J23102 and pSB3K3 Ligation of saturday, 10/11/2008 (wrong vector-insert ratio): 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight at 37°C
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-Receiver-Colicin E1
- Inoculation of Col E1 second mutagenesis step in 10 ml LB-amp; 5 h -> 37 °C
- Colonies 1-8 of Col E1 (5)
- Colonies 10-17 of Col E1 (1)
- Miniprep of liquidcultures (Col E1 (1.14;1.16;5.1;5.7): eluted in 35 µl H2O (Qiagen Miniprepkit)
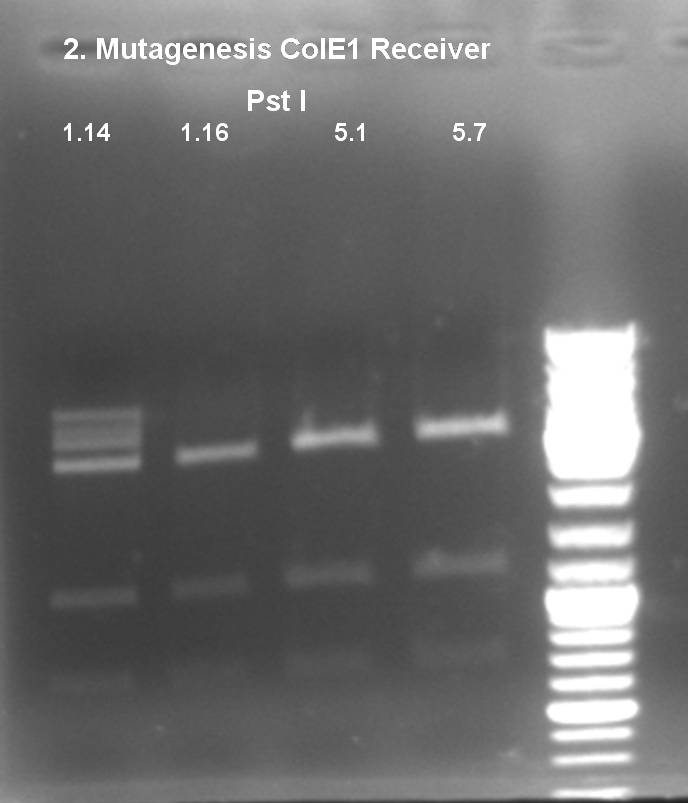
- Controlldigestion of second mutagenesis of Colicin E1 - Receiver with PstI: 2 h -> 37 °C
10.0 µl DNA (Miniprep) 2.0 µl BSA 10x (NEB) 0.5 µl PstI(NEB) 2.0 µl Buffer EcoRI 5.5 µl H2O ------- 20.0 µl
- Gel of Digestion: 1% Agarose, 135 V, 30 min
- Gelresults: It can not be concluded weather the mutation worked. The fragments that are expected with and without the mutated PstI-Site are very similar.
- Third Mutagenesis PCR:
5.0 µl pfu buffer 1.0 µl forward primer (10 µM Col E1_mut_Pst_2_fw) 1.0 µl reverse primer (10 µM Col E1_mut_Pst_2_rv) 1.0 µl dNTPs (12.5 mM, 50x) 0.5 µl template DNA - 5 to 50 ng plasmid DNA 40.5 µl milliQ water 1.0 µl turbo pfu polymerase (Stratagene) ---- 50.0 µl
95 °C 30 sec 95 °C 30 sec | 55 °C 60 sec | 16 cycles 68 °C 11 min | 4 °C constant
- Digestion with DpnI (NEB) of the third mutagenesis to cut parental plasmid: Added 1 µl of DpnI to the finished Mutagenesis PCR. DpnI cuts at methylated GATC sites, so that only parental plasmids without the mutation are cutted. 2 h 35 min -> 37 °C
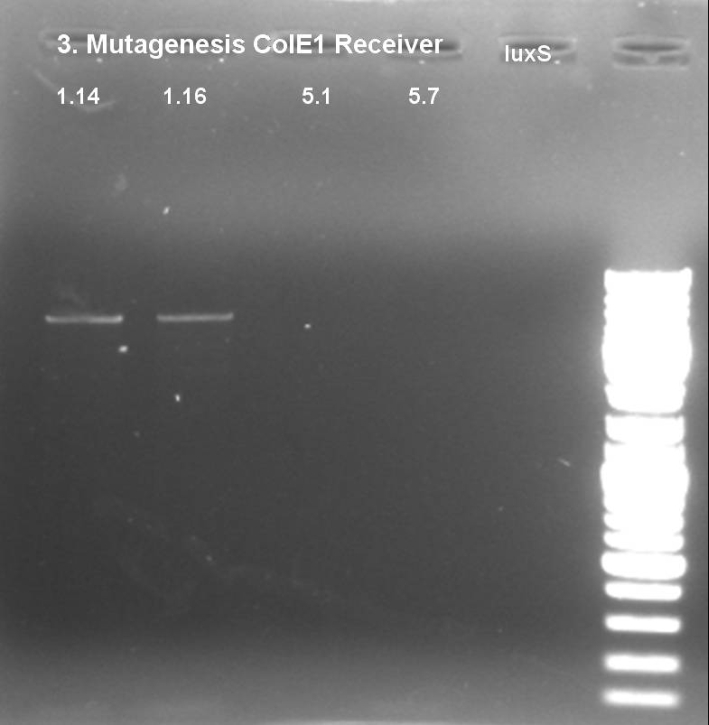
- Controlgel of third Mutagenesis (PstI_2): 1 % Agarose, 135 V, 30 min; DNA only visible in products from the colonie 1 (1.14 and 1.16) -> probably the mutagenesis worked on this plasmid.
- PCR Purification Kit (Qiagen) to purify the plasmids. Eluated in 35 µl H2O
- Transformation of prior mutagenesis: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight
Standardization of pSB1A2-colE9plasmid-Receiver
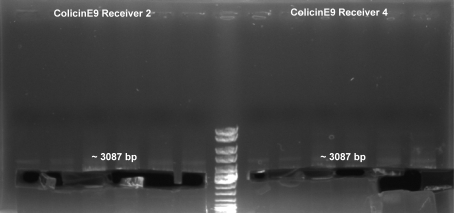
- PCR to add right BioBrick prefix and suffix to mutated ColicinE9-Receiver (without EcoRI site, colonies 2 & 4)
2.5 µl Primer fw T9002 (10 µM) 2.5 µl Primer rv (10 µM) 18.0 µl H2O 25.0 µl Phusion MasterMix (Finnzymes, NEB) 2.0 µl Template (ColE9-Receiver mutated) ------- 50.0 µl
98 °C 30 sec 98 °C 10 sec | 56 °C 30 sec | 25 cycles 72 °C 1 min 30 sec | 72 °C 10 min 4 °C constant
- Gel to purify PCR products: 0.7%, 135 V, 30 min
- Gelextraction: eluted in 35 µl H2O (Qiagen, Gelextraction Kit)
- Digestion of ColE9-Receiver PCR product with EcoRI and SpeI: 2h -> 37 °C
6.5 µl H2O 5.0 µl BSA 10x (NEB) 5.0 µl EcoRI Buffer (NEB) 0.5 µl EcoRI (NEB) 1.0 µl SpeI (NEB) 32.0 µl DNA ------- 50.0 µl
- Ligation of ColicinE9-Receiver (BioBrick) with pSB1A2 (3:1): 1h -> 22 °C, 20 min 65 °C
ColicinE9-Receiver (4) 1.1 µl pSB1A2 14.9 µl ColicinE9-Receiver 2.0 µl T4 DNA Ligase 2.0 µl T4 DNA Ligase Buffer ------- 20.0 µl
ColicinE9-Receiver (2) 1.8 µl pSB1A2 14.2 µl ColicinE9-Receiver 2.0 µl T4 DNA Ligase 2.0 µl T4 DNA Ligase Buffer ------- 20.0 µl
- Transformation of prior ligation: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight
[back]
 "
"