Team:USTC/Component
From 2008.igem.org
(→1. K082035) |
(→Differetiation Components) |
||
| (23 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
|} | |} | ||
| - | {| style="color:#FFFFFF;background-color:#66B3FF;" cellpadding="1" cellspacing="2" border="3" bordercolor="#555" width=" | + | {| style="color:#FFFFFF;background-color:#66B3FF;" cellpadding="1" cellspacing="2" border="3" bordercolor="#555" width="870px" align="center" |
|- | |- | ||
!align="center"|[[Team:USTC|Home]] | !align="center"|[[Team:USTC|Home]] | ||
| Line 23: | Line 23: | ||
!align="center"|[[Team:USTC/Project|The Project]] | !align="center"|[[Team:USTC/Project|The Project]] | ||
!align="center"|[[Team:USTC/Component|Components]] | !align="center"|[[Team:USTC/Component|Components]] | ||
| + | !align="center"|[[Team:USTC/Results|Results]] | ||
!align="center"|[[Team:USTC/Parts|Parts Submitted to the Registry]] | !align="center"|[[Team:USTC/Parts|Parts Submitted to the Registry]] | ||
!align="center"|[[Team:USTC/Notebook|Notebook]] | !align="center"|[[Team:USTC/Notebook|Notebook]] | ||
| Line 28: | Line 29: | ||
<br><br><br> | <br><br><br> | ||
| - | == | + | ==Differetiation Components== |
<br> | <br> | ||
| - | We design a time-dependent gene regulation system to construct a self-organized system. The | + | We design a time-dependent gene regulation system to construct a self-organized system. The RhlI/RhlR system is employed to report the cell-population density. The sender cell will appear first when the density reaches a certain level. And it will tell other cells around it to be the receiver cells by sending the ligands into the environment. After receiving the ligands, the Cre recombinase will be expressed to "scissor" the function of sending ligands. In this way, a positional differentiation will be achieved. |
| + | |||
<br> | <br> | ||
| + | |||
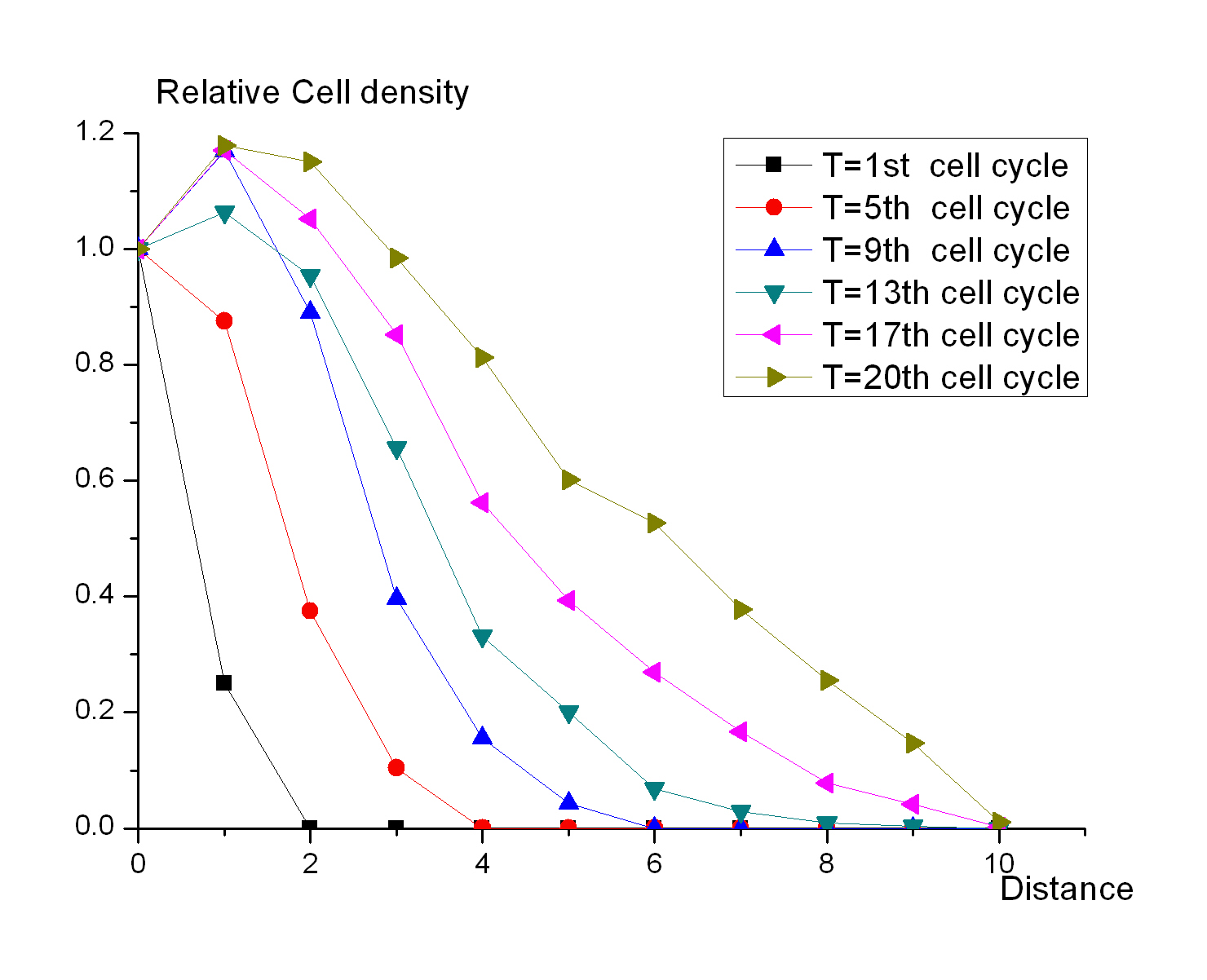
===The cell density of the colony on the plate=== | ===The cell density of the colony on the plate=== | ||
[[Image:USTC_cell_density.jpg|right|thumb|200px|Fig 1 .The changes of cell relative density related to the distance at different time points]] | [[Image:USTC_cell_density.jpg|right|thumb|200px|Fig 1 .The changes of cell relative density related to the distance at different time points]] | ||
<br> | <br> | ||
| - | Cell Automata is employed to mimic the states of a colony growing on the plate. The first seed is placed on the center of the table. Each seed has a child in the next generation, and the child seed is randomly putted into the grid next to its parent. It is the simplest simulation, but it can suggest how the bacteria grow on the plate when the resources are affluent. Fig 1 shows the changes of cell relative density related to | + | Cell Automata is employed to mimic the states of a colony growing on the plate. The first seed is placed on the center of the table. Each seed has a child in the next generation, and the child seed is randomly putted into the grid next to its parent. It is the simplest simulation, but it can suggest how the bacteria grow on the plate when the resources are affluent. Fig 1 shows the changes of cell relative density related to distance at different time points.And the result shows that 1) the colony can be divided into two parts, the central region and the peripheral region, according to the density of cells; 2) the central region will expand during the growth of the colony; 3) the difference between the peripheral region and the central region is evident enough to be sensed by the concentration of signal molecules. |
<br> | <br> | ||
<br> | <br> | ||
===Quorum Sensing System=== | ===Quorum Sensing System=== | ||
| - | RhlI/RhlR, one type of quorum sensing systems, is | + | RhlI/RhlR, one type of quorum sensing systems, is used as messengers to inform the cell of the local environment where it lives. RhlI is the enzyme that catalyzes the synthesis of C4HSL and RhlR is the receptor of C4HSL. If the concentration of C4HSL reaches a certain level, the RhlR will elevate the expression level of downstream genes of RhlR promoter to a great extent, which marks the initiation of differentiation. There are three genes downstream: RhlI, LuxI and LacI. RhlI can upregulate the expression of downstream genes by forming a positive feedback cycle. The LuxI gene is transcribed to synthesize the signal molecule 3OC6HSL, sending the differentiation order to other cells. However, LacI acts against AHL inhibiting the expression of differentiation executor. |
| - | In order to build the system, we have designed and constructed several protein-coding units | + | In order to build up the system, we have designed and constructed several protein-coding units |
| - | as follows | + | as follows, and more will be done in the next step. |
| + | |||
====1. K082035==== | ====1. K082035==== | ||
[[Image:USTC_RhlI_generator.jpg]] | [[Image:USTC_RhlI_generator.jpg]] | ||
<br> | <br> | ||
| - | In this sequence, we ligated the ptetR, RBS, RhlI | + | In this sequence, we ligated the ptetR, RBS, RhlI and terminator together, so as to produce the C4HSL constitutively. The sequencing result and the electrophoresis both demonstrate that the sequence of this unit is correct. Moreover, although it is too difficult for us to directly test whether RhlI can generate C4HSL as we could not obtain the purified C4HSL molecule, we constructed another sequence comprised by ptetR, RBS, RhlI, RBS, GFP and terminator. The green fluorescence we have observed confirmed the normal expression of RhlI indirectly. |
==== 2. K082021==== | ==== 2. K082021==== | ||
| + | [[Image:USTC_RhlR_generator.jpg]] | ||
| + | <br> | ||
The sequence codes for RhlR protein and the protein is constitutively expressed. | The sequence codes for RhlR protein and the protein is constitutively expressed. | ||
==== 3. K082020==== | ==== 3. K082020==== | ||
| - | Similarly, the sequence expresses the | + | [[Image:USTC_LuxR_generator.jpg]] |
| + | <br> | ||
| + | Similarly, the sequence expresses the LuxR protein constitutively. | ||
==== 4. K082045==== | ==== 4. K082045==== | ||
| - | This is the initiation part of differentiation in our project. The sequence codes for RhlI, LacI and LuxI and expresses them at a high level only when there are both RhlR and | + | [[Image:USTC_initiator.jpg]] |
| - | ==== | + | <br> |
| - | In our project, we need to use two types of signal transduction systems, and we choose to utilize the RhlI/RhlR and LuxI/LuxR systems. Our selection is based on the proof that the two kinds of systems do not | + | This is the initiation part of differentiation in our project. The sequence codes for RhlI, LacI and LuxI and expresses them at a high level only when there are both RhlR and C4HSL present. In our design, we hope the concentration of C4HSL increases to a certain amount but not too much, so a weaker RBS is used. Moreover, the proteins Lac repressor and AHL are also generated. |
| + | |||
| + | ====Cross-talk==== | ||
| + | In our project, we need to use two types of signal transduction systems, and we choose to utilize the RhlI/RhlR and LuxI/LuxR systems. Our selection is based on the proof (reference #10) that the two kinds of systems do not exchange the acyl-homoserine lactone signals. | ||
<br> | <br> | ||
| + | |||
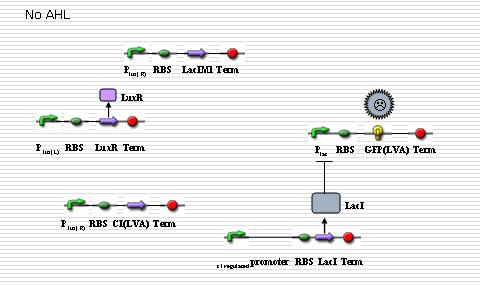
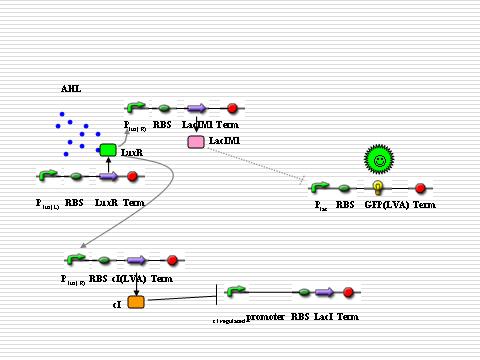
===NOR gate && Cre recombinase=== | ===NOR gate && Cre recombinase=== | ||
====NOR Gate==== | ====NOR Gate==== | ||
| - | The executor part of differentiation is the Cre recombinase that is regulated by | + | The executor part of differentiation is the Cre recombinase that is regulated by the NOR gate. The designed NOR gate is regulated by two proteins: LacI and LuxR. NOR gate will only be turned on with AHL present and LacI absent. Here the logical gate is utilized to distinguish whether the signal molecule is endogenic or ectogenic. At the beginning, the expression will be inhibited by LuxR. Once LuxI is expressed, AHL will be synthesized and spread throughout the colony. If AHL is endogenic, LacI will be expressed simultaneously so that the executor is not activated. But if AHL is ectogenic, before the concentration of LacI in the cell reaches a sufficient level, the NOR gate is turned on and the executor will be expressed, leading to the shutting off of the AHL generator. |
{|align="center" border="1" | {|align="center" border="1" | ||
| Line 93: | Line 105: | ||
{|align=center | {|align=center | ||
| - | |[[Image:USTC_rec_3.jpg| | + | |[[Image:USTC_rec_3.jpg|400px|thumb|Fig 2.General recombine system]] |
|width=30px| | |width=30px| | ||
| - | |[[Image:USTC_rec_4.jpg| | + | |[[Image:USTC_rec_4.jpg|350px|thumb|Fig 3.The overview of the whole system]] |
| | | | ||
|} | |} | ||
<br> | <br> | ||
| - | == Report | + | == Report Components== |
The response to distance requires that the reporter gene only responds to a narrow range of signal concentrations, which means the reporter gene will not be expressed when the concentration of signals is either too high or too low. There are some different ways to deal with the problem: | The response to distance requires that the reporter gene only responds to a narrow range of signal concentrations, which means the reporter gene will not be expressed when the concentration of signals is either too high or too low. There are some different ways to deal with the problem: | ||
| Line 108: | Line 120: | ||
In “sender” cells, the tetR promoter expresses LuxI constitutively so that an AHL gradient is built up around the senders. When the AHL concentration reaches a certain level, the LuxR promoter is activated to a large extent so that LacIM1 protein (a mutant of LacI) and cI protein will be expressed in a large amount. The cI protein will shut off the expression of LacI while the LacIM1 protein will inhibit the expression of GFP regulated by LacR promoter. When the AHL concentration is intermediate, the cI protein is still capable to stop the LacI expression while the LacIM1 protein can not inactivate the LacR promoter any longer, so a certain level of GFP can be observed. On the other side, when the AHL concentration is too low, neither cI nor LacIM1 will be expressed. Without cI protein, the lambda promoter will be activated and LacI protein is produced, which inactivates the LacR promoter so that GFP could be barely observed. All the above are shown as the figures below. | In “sender” cells, the tetR promoter expresses LuxI constitutively so that an AHL gradient is built up around the senders. When the AHL concentration reaches a certain level, the LuxR promoter is activated to a large extent so that LacIM1 protein (a mutant of LacI) and cI protein will be expressed in a large amount. The cI protein will shut off the expression of LacI while the LacIM1 protein will inhibit the expression of GFP regulated by LacR promoter. When the AHL concentration is intermediate, the cI protein is still capable to stop the LacI expression while the LacIM1 protein can not inactivate the LacR promoter any longer, so a certain level of GFP can be observed. On the other side, when the AHL concentration is too low, neither cI nor LacIM1 will be expressed. Without cI protein, the lambda promoter will be activated and LacI protein is produced, which inactivates the LacR promoter so that GFP could be barely observed. All the above are shown as the figures below. | ||
| - | [[Image:USTC_exp_1.jpg|left|250px|thumb|Fig 4.The low concerntration condition]] | + | ::[[Image:USTC_exp_1.jpg|left|250px|thumb|Fig 4.The low concerntration condition]] |
[[Image:USTC_exp_2.jpg|left|250px|thumb|Fig 5.The medium concerntration condition]] | [[Image:USTC_exp_2.jpg|left|250px|thumb|Fig 5.The medium concerntration condition]] | ||
[[Image:USTC_exp_3.jpg|left|250px|thumb|Fig 6.The high concerntration condition]] | [[Image:USTC_exp_3.jpg|left|250px|thumb|Fig 6.The high concerntration condition]] | ||
<br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| - | + | ||
| - | + | To organize this part to our system, we should first standard all the genes that don’t include in biobrick parts. We use PCR to clone the genes from plasmids given as gifts fromRon Weiss lab. PCR is also used to mutant the biobrick restriction sites follow the [https://2008.igem.org/USTC/Notebook/Point_mutation_Quick-Change_method Quick-Change method] described below. | |
| + | we’ve already cloned GFP(LVA)(GFP with LVA tag [http://nobelprize.org/nobel_prizes/chemistry/laureates/2008/ GFP GOT THIS YEAR'S NOBEL PRIZE!]), LuxR-G2F(a mutant of LuxR), LacI,LacIM1(a mutant of LacI with low affinity to lac promoter), some of which have one or more point mutations to avoid the standard restriction sites. after putting parts such as RBS, promoter and terminator together, we have made some of the protein-coding genes work successfully, e.g. GFP(LVA) under the control of lac promoter lights once stimulated.[https://2008.igem.org/Team:USTC/Results (See our Results) ]Unfortunately, time is not enough for us to put all the units together and test their functions. | ||
===XOR GATE=== | ===XOR GATE=== | ||
| Line 137: | Line 150: | ||
|- | |- | ||
|} | |} | ||
| - | In another way , the response in a narrow range can be | + | In another way, the response in a narrow range can be achieved by an XOR gate. The promoter is activated when only one protein binds to the DNA. The downstream genes can not be expressed either when two proteins bind or don't bind the operator. The truth table is shown on the right.And here is a interesting pheonomenon that we can utilize: if we placed pLuxR in the upstream of -35 site, the downstream genes will be transcribed when AHL is present, and if we placed pLuxR between -35 and -10, the downstream genes will be inactivated by AHL. So we can employ two similar proteins that respond to concentration distinctly to work as an XOR gate. The two proteins will not respond both at low concentrations and at the high concentrations. Only in intermediate concentrations, the protein with higher sensitivity will bind DNA and express the downstream genes. |
| - | + | ||
| - | + | ||
| + | ===Computational Protein Design=== | ||
| + | How to change protein's sensitivity to ligands? Computational protein design is one easier way. From Protein Data Bank, we can obtain the structure of LuxR and AHL. The analysis tells us the contribution of each residue to docking of the whole protein. So in silico screen we can get the answers about what changes can weaken or strengthen the binding capability. | ||
| + | |||
| + | <br> | ||
==References== | ==References== | ||
---- | ---- | ||
| Line 151: | Line 166: | ||
*4. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora | *4. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora | ||
:Yi-Hu Dong, Jin-Ling Xu, Xian-Zhen Li, and Lian-Hui Zhang* PNAS March 28, 2000, 3526–3531 | :Yi-Hu Dong, Jin-Ling Xu, Xian-Zhen Li, and Lian-Hui Zhang* PNAS March 28, 2000, 3526–3531 | ||
| + | *5. Computational redesign of endonuclease DNA binding and cleavage specificity. | ||
| + | :Ashworth, J., Havranek, J.J., Duarte, C.M., Sussman, D., Monnat, R.J., Stoddard, B.L. & Baker, D.Nature, 2006, Vol. 441(7093), pp. 656-659 | ||
| + | *6. Improved Pruning algorithms and Divide-and-Conquer strategies for Dead-End Elimination, with application to protein design | ||
| + | :Georgiev, I., Lilien, R.H. & Donald, B.R. Bioinformatics (Oxford, England), 2006, Vol. 22, pp. e174-83 | ||
| + | *7. Computing van der Waals energies in the context of the rotamer approximation. | ||
| + | :Grigoryan, G., Ochoa, A. & Keating, A.E. Proteins, 2007, Vol. 68(4), pp. 863-878 | ||
| + | *8. Progress in computational protein design. | ||
| + | :Lippow, S.M. & Tidor, B. Curr Opin Biotechnol, 2007, Vol. 18(4), pp. 305-311 | ||
| + | *9. New algorithms and an in silico benchmark for computational enzyme design. | ||
| + | :Zanghellini, A., Jiang, L., Wollacott, A.M., Cheng, G., Meiler, J., Althoff, E.A., R?thlisberger, D. & Baker, D.Protein Sci, 2006, Vol. 15(12), pp. 2785-2794 | ||
| + | *10. Directed Evolution of Vibrio fischeri LuxR for Improved Response to Butanoyl-Homoserine Lactone. | ||
| + | :Andrew C. Hawkins, Frances H. Arnold, Rainer Stuermer,Bernhard Hauer, and Jared R. Leadbetter1*, 2007, APPLIED AND ENVIRONMENTAL MICROBIOLOGY, p. 5775–5781. | ||
Latest revision as of 03:20, 30 October 2008
| Home | The Team | The Project | Components | Results | Parts Submitted to the Registry | Notebook |
|---|
Contents |
Differetiation Components
We design a time-dependent gene regulation system to construct a self-organized system. The RhlI/RhlR system is employed to report the cell-population density. The sender cell will appear first when the density reaches a certain level. And it will tell other cells around it to be the receiver cells by sending the ligands into the environment. After receiving the ligands, the Cre recombinase will be expressed to "scissor" the function of sending ligands. In this way, a positional differentiation will be achieved.
The cell density of the colony on the plate
Cell Automata is employed to mimic the states of a colony growing on the plate. The first seed is placed on the center of the table. Each seed has a child in the next generation, and the child seed is randomly putted into the grid next to its parent. It is the simplest simulation, but it can suggest how the bacteria grow on the plate when the resources are affluent. Fig 1 shows the changes of cell relative density related to distance at different time points.And the result shows that 1) the colony can be divided into two parts, the central region and the peripheral region, according to the density of cells; 2) the central region will expand during the growth of the colony; 3) the difference between the peripheral region and the central region is evident enough to be sensed by the concentration of signal molecules.
Quorum Sensing System
RhlI/RhlR, one type of quorum sensing systems, is used as messengers to inform the cell of the local environment where it lives. RhlI is the enzyme that catalyzes the synthesis of C4HSL and RhlR is the receptor of C4HSL. If the concentration of C4HSL reaches a certain level, the RhlR will elevate the expression level of downstream genes of RhlR promoter to a great extent, which marks the initiation of differentiation. There are three genes downstream: RhlI, LuxI and LacI. RhlI can upregulate the expression of downstream genes by forming a positive feedback cycle. The LuxI gene is transcribed to synthesize the signal molecule 3OC6HSL, sending the differentiation order to other cells. However, LacI acts against AHL inhibiting the expression of differentiation executor. In order to build up the system, we have designed and constructed several protein-coding units as follows, and more will be done in the next step.
1. K082035
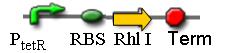
In this sequence, we ligated the ptetR, RBS, RhlI and terminator together, so as to produce the C4HSL constitutively. The sequencing result and the electrophoresis both demonstrate that the sequence of this unit is correct. Moreover, although it is too difficult for us to directly test whether RhlI can generate C4HSL as we could not obtain the purified C4HSL molecule, we constructed another sequence comprised by ptetR, RBS, RhlI, RBS, GFP and terminator. The green fluorescence we have observed confirmed the normal expression of RhlI indirectly.
2. K082021
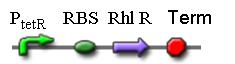
The sequence codes for RhlR protein and the protein is constitutively expressed.
3. K082020
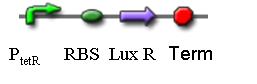
Similarly, the sequence expresses the LuxR protein constitutively.
4. K082045
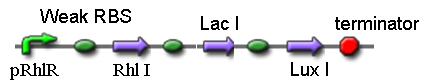
This is the initiation part of differentiation in our project. The sequence codes for RhlI, LacI and LuxI and expresses them at a high level only when there are both RhlR and C4HSL present. In our design, we hope the concentration of C4HSL increases to a certain amount but not too much, so a weaker RBS is used. Moreover, the proteins Lac repressor and AHL are also generated.
Cross-talk
In our project, we need to use two types of signal transduction systems, and we choose to utilize the RhlI/RhlR and LuxI/LuxR systems. Our selection is based on the proof (reference #10) that the two kinds of systems do not exchange the acyl-homoserine lactone signals.
NOR gate && Cre recombinase
NOR Gate
The executor part of differentiation is the Cre recombinase that is regulated by the NOR gate. The designed NOR gate is regulated by two proteins: LacI and LuxR. NOR gate will only be turned on with AHL present and LacI absent. Here the logical gate is utilized to distinguish whether the signal molecule is endogenic or ectogenic. At the beginning, the expression will be inhibited by LuxR. Once LuxI is expressed, AHL will be synthesized and spread throughout the colony. If AHL is endogenic, LacI will be expressed simultaneously so that the executor is not activated. But if AHL is ectogenic, before the concentration of LacI in the cell reaches a sufficient level, the NOR gate is turned on and the executor will be expressed, leading to the shutting off of the AHL generator.
| LuxR | LacI | Exp |
| 0 | 0 | 1 |
| 1 | 0 | 0 |
| 0 | 1 | 0 |
| 1 | 1 | 0 |
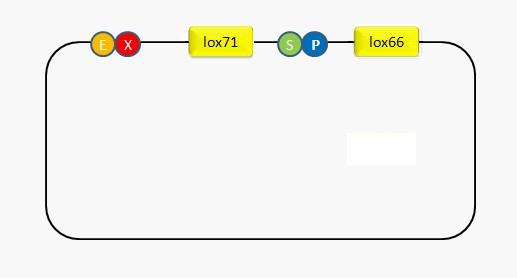
Recombinase
In order to utilize the recombination system repetitively, we design an adaptor where we can put in any part that needs recombining. To accomplish this goal, the restriction sites of SpeI and PstI are placed between lox66 and lox71. In this way, any fragment digested by XbaI and PstI can be inserted between the two recombination sites. We locate the recombination sites into the blank plasmid pSB3C5. The sequencing result as well as the insertion of GFP proves that the system can work successfully.(Fig2,Fig3)
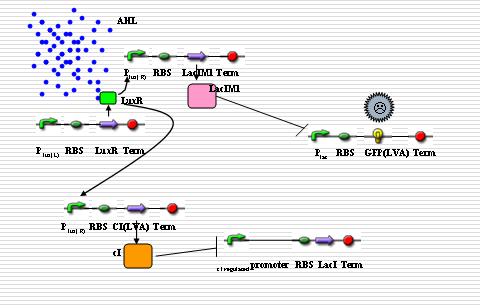
Report Components
The response to distance requires that the reporter gene only responds to a narrow range of signal concentrations, which means the reporter gene will not be expressed when the concentration of signals is either too high or too low. There are some different ways to deal with the problem:
Multi-regulatory system
The idea comes from the Nature letter “A Synthetic Multicellular System for Programmed Pattern Formation”. In this paper, based on quorum sensing, the engineered ‘receiver’ cells are programmed to form ring-like patterns while the chemical gradient of the acyl-homoserine lactone signal synthesized by ‘sender’ cells is formed. The ‘sender’ and ‘receiver’ cells can be organized to our system.
In “sender” cells, the tetR promoter expresses LuxI constitutively so that an AHL gradient is built up around the senders. When the AHL concentration reaches a certain level, the LuxR promoter is activated to a large extent so that LacIM1 protein (a mutant of LacI) and cI protein will be expressed in a large amount. The cI protein will shut off the expression of LacI while the LacIM1 protein will inhibit the expression of GFP regulated by LacR promoter. When the AHL concentration is intermediate, the cI protein is still capable to stop the LacI expression while the LacIM1 protein can not inactivate the LacR promoter any longer, so a certain level of GFP can be observed. On the other side, when the AHL concentration is too low, neither cI nor LacIM1 will be expressed. Without cI protein, the lambda promoter will be activated and LacI protein is produced, which inactivates the LacR promoter so that GFP could be barely observed. All the above are shown as the figures below.
To organize this part to our system, we should first standard all the genes that don’t include in biobrick parts. We use PCR to clone the genes from plasmids given as gifts fromRon Weiss lab. PCR is also used to mutant the biobrick restriction sites follow the Quick-Change method described below. we’ve already cloned GFP(LVA)(GFP with LVA tag [http://nobelprize.org/nobel_prizes/chemistry/laureates/2008/ GFP GOT THIS YEAR'S NOBEL PRIZE!]), LuxR-G2F(a mutant of LuxR), LacI,LacIM1(a mutant of LacI with low affinity to lac promoter), some of which have one or more point mutations to avoid the standard restriction sites. after putting parts such as RBS, promoter and terminator together, we have made some of the protein-coding genes work successfully, e.g. GFP(LVA) under the control of lac promoter lights once stimulated.(See our Results) Unfortunately, time is not enough for us to put all the units together and test their functions.
XOR GATE
| LuxR-M1 | LuxR-M2 | Exp |
| 0 | 0 | 0 |
| 1 | 0 | 1 |
| 1 | 1 | 0 |
In another way, the response in a narrow range can be achieved by an XOR gate. The promoter is activated when only one protein binds to the DNA. The downstream genes can not be expressed either when two proteins bind or don't bind the operator. The truth table is shown on the right.And here is a interesting pheonomenon that we can utilize: if we placed pLuxR in the upstream of -35 site, the downstream genes will be transcribed when AHL is present, and if we placed pLuxR between -35 and -10, the downstream genes will be inactivated by AHL. So we can employ two similar proteins that respond to concentration distinctly to work as an XOR gate. The two proteins will not respond both at low concentrations and at the high concentrations. Only in intermediate concentrations, the protein with higher sensitivity will bind DNA and express the downstream genes.
Computational Protein Design
How to change protein's sensitivity to ligands? Computational protein design is one easier way. From Protein Data Bank, we can obtain the structure of LuxR and AHL. The analysis tells us the contribution of each residue to docking of the whole protein. So in silico screen we can get the answers about what changes can weaken or strengthen the binding capability.
References
- 1.A Synthetic Multicellular System for Programmed Pattern Formation
- Subhayu Basu, Yoram Gerchman, Cynthia H. Collins, Frances H. Arnold & Ron Weiss NATURE April 28, 2005, 1130-1134
- 2.Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples
- Anjali Kumari & Patrizia Pasini & Sylvia Daunert Anal Bioanal Chem (2008) 391:1619–1627
- 3.Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: A signaling mechanism involved in associations with higher organisms
- Matthew R. Parsek and E. Peter Greenberg PNAS August 1, 2000, 8789–8793
- 4. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora
- Yi-Hu Dong, Jin-Ling Xu, Xian-Zhen Li, and Lian-Hui Zhang* PNAS March 28, 2000, 3526–3531
- 5. Computational redesign of endonuclease DNA binding and cleavage specificity.
- Ashworth, J., Havranek, J.J., Duarte, C.M., Sussman, D., Monnat, R.J., Stoddard, B.L. & Baker, D.Nature, 2006, Vol. 441(7093), pp. 656-659
- 6. Improved Pruning algorithms and Divide-and-Conquer strategies for Dead-End Elimination, with application to protein design
- Georgiev, I., Lilien, R.H. & Donald, B.R. Bioinformatics (Oxford, England), 2006, Vol. 22, pp. e174-83
- 7. Computing van der Waals energies in the context of the rotamer approximation.
- Grigoryan, G., Ochoa, A. & Keating, A.E. Proteins, 2007, Vol. 68(4), pp. 863-878
- 8. Progress in computational protein design.
- Lippow, S.M. & Tidor, B. Curr Opin Biotechnol, 2007, Vol. 18(4), pp. 305-311
- 9. New algorithms and an in silico benchmark for computational enzyme design.
- Zanghellini, A., Jiang, L., Wollacott, A.M., Cheng, G., Meiler, J., Althoff, E.A., R?thlisberger, D. & Baker, D.Protein Sci, 2006, Vol. 15(12), pp. 2785-2794
- 10. Directed Evolution of Vibrio fischeri LuxR for Improved Response to Butanoyl-Homoserine Lactone.
- Andrew C. Hawkins, Frances H. Arnold, Rainer Stuermer,Bernhard Hauer, and Jared R. Leadbetter1*, 2007, APPLIED AND ENVIRONMENTAL MICROBIOLOGY, p. 5775–5781.
 "
"