Team:Heidelberg/Notebook/Killing I/Notebook/week10
From 2008.igem.org
(→Characterization of oriT) |
(→Characterize of oriT) |
||
| (2 intermediate revisions not shown) | |||
| Line 707: | Line 707: | ||
**pcr was not successful, gel included no pcr product | **pcr was not successful, gel included no pcr product | ||
| - | + | === Characterize of oriT=== | |
| + | *Quantitatively test for oriT | ||
| + | Donor: overnight culture Cotransformation Top10 J01103+pUB307(12) OD(600nm): 2.844<br> | ||
| + | Recipient: overnight culture Top10 pBAD 33 OD(600nm): 3.346<br> | ||
| + | **Centrifuge 250ul overnight culture in 1.5ml eppi for 2min at 13000rpm, 10samples donor, 10samples recipient | ||
| + | **Wash the pellet twice with LB medium | ||
| + | **Resolve the pellet in 250ul LB medium | ||
| + | **Centrifuge the washed recipient for 2min at 13000rpm, discard the fluid | ||
| + | **Add the washed donor suspension | ||
| + | **Vortex and resolve the pellet | ||
| + | **Centrifuge the mix for 1min at 13000rpm | ||
| + | **Resolve the pellet in 100ul LB | ||
| + | **Put membrane filter on the LB agar | ||
| + | **Pipett the suspension on membrane filter (10samples) | ||
| + | **Incubate the plates with membrane filter at 37°C | ||
| + | **Put directly one membrane filter into 1ml LB in an 1.5ml eppi | ||
| + | **Vortex the eppi for 30sec, dilute for 10-4 and 10-5, plate them out on LB/Amp+Cm plates (0min) | ||
| + | **After 6, 12, 18, 24, 30, 36, 42, 48, 54min repeat the last two steps.<br> | ||
| + | **Negative control plates: | ||
| + | ***LB/Cm+Amp: | ||
| + | 100ul donor overnight culture<br> | ||
| + | 100ul recipient overnight culture<br> | ||
| + | **Cell number determination | ||
| + | ***LB/Cm: 100ul 10-6 recipient overnight culture | ||
| + | ***LB/Kan+Amp: 100ul 10-6 donor overnight culture | ||
| + | <br> | ||
| + | *Result: | ||
| + | **Negative control: negative | ||
| + | **Colony on LB/Cm: 324 (Titer of recipient: 3.24e9/ml) | ||
| + | **Colony on LB/Kan+Amp: 373 (Titer of donor: 3.73e9/ml) | ||
| + | **Colony on other LB/Cm+Amp plates: | ||
| + | **10-4 dilute: impossible for counting | ||
| + | **10-5 dilute:<br> | ||
| + | {| class="wikitable" | ||
| + | |- bgcolor=grey | ||
| + | ! height=20px. width=200px | Time || width=250px | Colony | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |0 || 150 | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |6 || 500 | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |12 || 780 | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |18 || 1160 | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |24 || 1400 | ||
| + | |-align="center" | ||
| + | | style="font-weight:bold;" |30 || 3360 | ||
| + | |- | ||
| + | |} | ||
| + | ***36min to 54 min: impossible for counting | ||
| + | <br> | ||
| + | *Make glycerol stock for Cotransformation Top10 oriT+pUB307 | ||
| + | **1ml overnight culture of Top10 oriT+pUB307 (12)+ 150ul 80% glycerol | ||
| + | **Vortex | ||
| + | **1hr RT | ||
| + | **Freeze at -80°C | ||
==Thursday, 10/09/08 == | ==Thursday, 10/09/08 == | ||
Latest revision as of 14:09, 29 October 2008


| << Week 9 | Overview | Week 11 >> |
|---|
Week 10
Contents |
Monday, 10/06/08
Cloning of CmR in pBluescript
Proceeding of the overnight ligations
- 8 transformation of the CmR overnight ligations
- 3 control transformation of Term1, CmR3 and pBluesript backbone
Proceeding of the minipreps from friday
- digestions of CmR in pBlue Miniprep to check for sequencing
- digestion with SacI, KpnI and EcoRI
- expected bands: 2859, 586, 272
- digestion with SacI, KpnI and EcoRI
- digestion with DraI
- expected bands: 1480, 1187, 692, 339, 19
- digestion with DraI
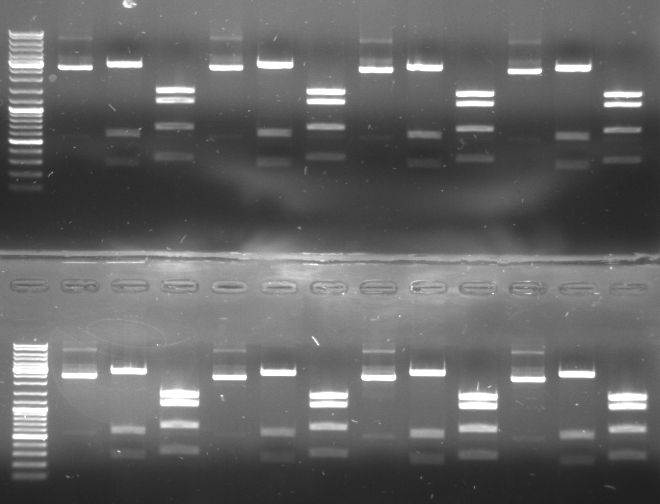
- Gel
- top:
- lane1-3: CmR1 (undigested, SacI/KpnI/EcoRI, DraI)
- lane4-6: CmR2
- lane7-10: CmR3a
- lane11-13: CmR3b
- bottom
- lane1-3: CmR4 (undigested, SacI/KpnI/EcoRI, DraI)
- lane4-6: CmR5
- lane7-10: CmR7
- lane11-13: CmR8
- according to the digestions we have the chloramphenicol resistance cassette in pBluescript
- Retrafo of CmR minipreps because we don't have enough DNA and unfortunately no glycerol stocks
- 3µl of CmR 1, 2, 3a, 3b
Cloning of CmR in standard plasmid
- PCR of CmR with CmR_prefix_fw and CmR_suffix_fw
25µl Phusion Master Mix 2x 1µl CmR_suffix_fw 1µl CmR_prefix_rev 1µl Maxiprep pBlue with insert (stock: ~200ng/µl, dilution: 1:20-->10ng) 22µl water ----- 50µl
PCR protocol 98°C 1min 98°C 10s | 61°C 10s | 26x 72°C 45s | 72°C 5min 4°C for ever
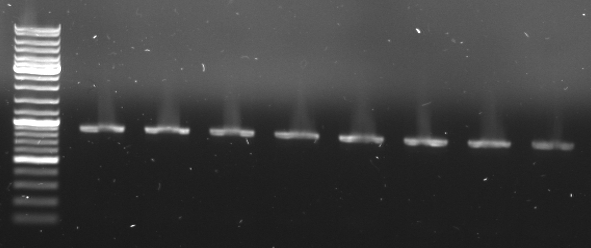
- Gel
- expected size: 851bp+43bp=894bp
- lane0:ladder
- lane1: CmR1
- lane2: CmR2
- lane3: CmR3a
- lane4: CmR3b
- lane5: CmR4
- lane6: CmR5
- lane7: CmR7
- lane8: CmR8
- cuted out CmR band and gel extraction kit
Cloning of cI in standard plasmid
- unwanted EcoRI restriction site in cI from geneart was mutated using standard protocol
- lane 1+2: mutated cI digested with EcoRI --> mutagenesis was succesfull
- maxiprep of this cI as well as J01003 in pSB1A2 were digested with EcoRI/PstI and XbaI/PstI to ligate this mutated cI in a standard plasmid (pSB1A2)
- lane 1-4: cI digested with EcoRI/PstI
- lane 5-9: cI digested with XbaI/PstI (lane 7 is ladder)
- lane 10+11: J01003 digested with XbaI/PstI
- lane 12+13: J01003 digested with EcoRI/PstI
--> digested cI and pSB1A2 backbone was cut out and ligated (15 min, RT) --> transformation in E.coli Top10 --> inoculation of liquid cultures --> miniprep --> send for sequencing and controll digest with EcoRI/PstI
- seqeuncing was succesfull
- digestion with EcoRI/PstI
- lane 1-6: mutated cI in pSB1A2 digested with EcoRI/PstI
results: sequencing and digestion pattern is correct
Characterization of oriT
- Inoculate the cells for conjugation test
- 5ml LB/chloramphenicol + glycerol stock Top10 pBAD 33
- 5ml LB/kanamycin/ampicilin + 1 colony Cotransformation Top10 J01103+pUB307(12)
- 5ml LB/kanamycin/ampicilin + 1 colony Cotransformation Top10 J01103+pUB307(22)
- 5ml LB/kanamycin/ampicilin + 1 colony Cotransformation Top10 J01103+pUB307(13)
- 5ml LB/kanamycin/ampicilin + 1 colony Cotransformation Top10 J01103+pUB307(23)
Tuesday, 10/07/08
Proceeding of cloning CmR in pBluescript
- a lot of single colonies of the transformation of the new ligations
- a lot of colonies on the retrafo
- inoculation of liquid culture of the 4 retrafos and the first 4 new ligations
- agar plates with new ligations are stored at 4 °C
Cloning of CmR in standard plasmid
- Digestion of CmR pcr product with PstI, XbaI
5µl NEB3 5µl BSA 10µl PCR product CmR (app. 50ng/µl (from gel) ) 1 µl PstI 1,5µl XbaI 32,5µl water
- 4 samples: 1,2, 3a, 3b
- 2h at 37°C
- PCR purification kit
- ligation of digested CmR pcr product in pSB1A2, 30min at room temperature
- transformation in TOP10, plated out on CmR plates
- ligation was done overnight as well
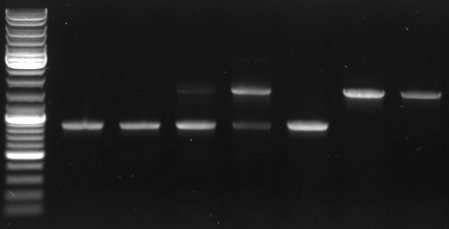
Phage cloning strategy two
- Digestion of KpnI mutagenesis PCR
3µl DNA 5µl NEB1 5µl BSA 1µl KpnI 1µl AgeI 36µl water
- 4 digestions with following mutagenesis pcr samples: 1,3,4,6
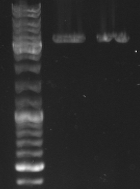
- Gel
- mutagensis pcr successful: 1690bp, 5050bp
- -->cut out 5050bp band
- mutagensis pcr unsuccessful: 1684bp, 1690bp, 3366bp
- lane0: DNA ladder mix
- lane1-3: digested pcr sample 1
- lane4-6: digested pcr sample 3
- lane7-9: digested pcr sample 4
- lane10-12: digested pcr sample 6
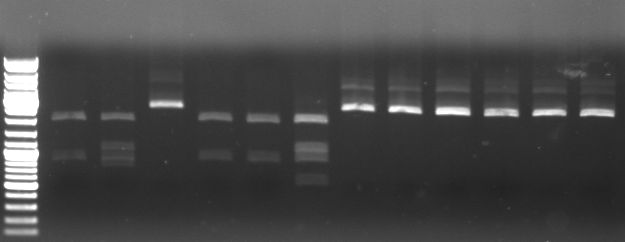
- Digestion of CmR out of CmR in pBluescript with KpnI SacI
5µl NEB1 5µl BSA 3µl CmR miniprep 1,5µl KpnI 1,5µl SacI 34µl water
--> digestion of CmR miniprep 1,2,4,5
- Gel
- expected: 858bp, 2859bp
- -->cut out 858bp band
- lane0:ladder
- lane1-3: digested miniprep 1
- lane4-6: digested miniprep 2
- lane7-9: digested miniprep 4
- lane10-12: digested miniprep 5
- ligation of CmR (KpnI/SacI), GFP (SacI/AgeI), pBluescript (KpnI/AgeI) 30min at room temperature
- transformation in TOP10
- ligation was done at 16°C over night as well
Characterization of oriT
- Qualitatively test for oriT
Donor: overnight culture Cotransformation Top10 oriT+pUB307(12), (22), (13), (23)
Recipient: overnight culture Top10 pBAD 33
- Centrifuge 500ul overnight culture in 1.5ml eppi for 2min at 13000rpm
- Wash the pellet twice with LB medium
- Resolve the pellet in 500ul LB medium
- Mix 500ul washed recipient cell suspension with 500ul washed donor cell suspension in 2ml eppi
- Vortex
- Incubate the mix at 37°C for 1hr
- Plates:
- LB/Cm: 100ul overnight culture Top10 pBAD 33
- LB/Kan+Amp:
100ul overnight culture Top10 oriT+pUB307(12)
100ul overnight culture Top10 oriT+pUB307(22)
100ul overnight culture Top10 oriT+pUB307(13)
100ul overnight culture Top10 oriT+pUB307(23)
- LB/Amp+Cm:
100ul overnight culture Top10 oriT+pUB307(12)
100ul overnight culture Top10 oriT+pUB307(22)
100ul overnight culture Top10 oriT+pUB307(13)
100ul overnight culture Top10 oriT+pUB307(23)
100ul overnight culture Top10 pBAD 33
100ul conjugation mix (12)
100ul conjugation mix (13)
100ul conjugation mix (22)
100ul conjugation mix (23)
- Result:
All LB/Cm positive; all LB/Kan+Amp positive; LB/Amp+Cm with donor or recipient negative; LB/Amp+Cm with conjugation mix positive -> like expectation
- Inoculate cells for conjugation test
- 5ml LB/chloramphenicol + 10ul overnight culture Top10 pBAD 33
- 5ml LB/kanamycin/ampicilin + 10ul overnight culture Cotransformation Top10 oriT+pUB307(12)
- 5ml LB/kanamycin/ampicilin + 10ul overnight culture Cotransformation Top10 oriT+pUB307(22)
- 5ml LB/kanamycin/ampicilin + 10ul overnight culture Cotransformation Top10 oriT+pUB307(13)
- 5ml LB/kanamycin/ampicilin + 10ul overnight culture Cotransformation Top10 oriT+pUB307(23)
Wednesday, 10/08/08
Phage cloning strategy two
- transformation of overnight ligations in TOP10
- screening pcr to check if ligation was successful using GFP_new_fw and GFP_new_rv
- pcr was not successful, gel included no pcr product
Characterize of oriT
- Quantitatively test for oriT
Donor: overnight culture Cotransformation Top10 J01103+pUB307(12) OD(600nm): 2.844
Recipient: overnight culture Top10 pBAD 33 OD(600nm): 3.346
- Centrifuge 250ul overnight culture in 1.5ml eppi for 2min at 13000rpm, 10samples donor, 10samples recipient
- Wash the pellet twice with LB medium
- Resolve the pellet in 250ul LB medium
- Centrifuge the washed recipient for 2min at 13000rpm, discard the fluid
- Add the washed donor suspension
- Vortex and resolve the pellet
- Centrifuge the mix for 1min at 13000rpm
- Resolve the pellet in 100ul LB
- Put membrane filter on the LB agar
- Pipett the suspension on membrane filter (10samples)
- Incubate the plates with membrane filter at 37°C
- Put directly one membrane filter into 1ml LB in an 1.5ml eppi
- Vortex the eppi for 30sec, dilute for 10-4 and 10-5, plate them out on LB/Amp+Cm plates (0min)
- After 6, 12, 18, 24, 30, 36, 42, 48, 54min repeat the last two steps.
- Negative control plates:
- LB/Cm+Amp:
100ul donor overnight culture
100ul recipient overnight culture
- Cell number determination
- LB/Cm: 100ul 10-6 recipient overnight culture
- LB/Kan+Amp: 100ul 10-6 donor overnight culture
- Cell number determination
- Result:
- Negative control: negative
- Colony on LB/Cm: 324 (Titer of recipient: 3.24e9/ml)
- Colony on LB/Kan+Amp: 373 (Titer of donor: 3.73e9/ml)
- Colony on other LB/Cm+Amp plates:
- 10-4 dilute: impossible for counting
- 10-5 dilute:
| Time | Colony |
|---|---|
| 0 | 150 |
| 6 | 500 |
| 12 | 780 |
| 18 | 1160 |
| 24 | 1400 |
| 30 | 3360 |
- 36min to 54 min: impossible for counting
- Make glycerol stock for Cotransformation Top10 oriT+pUB307
- 1ml overnight culture of Top10 oriT+pUB307 (12)+ 150ul 80% glycerol
- Vortex
- 1hr RT
- Freeze at -80°C
Thursday, 10/09/08
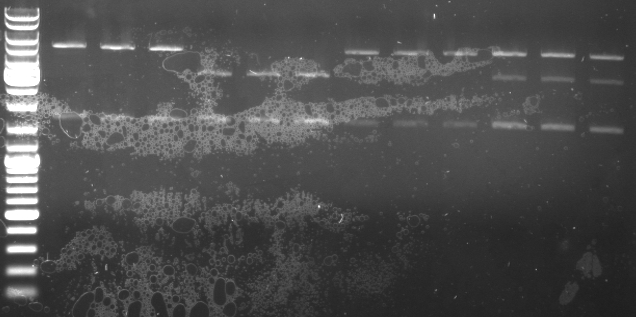
Cloning of CmR in standard plasmid
- miniprep of 12 from the 20 overnight cultures (CmR in pSB1A2)
- sample nr. 1.1-1.3, 2.1-2.3, 3.1-3.3, 4.1-4.3
- digestion of the 12 minipreps with EcoRI and PstI
4µl EcoRI buffer 4µl BSA 1µl EcoRI 1µl PstI 3µl Miniprep DNA 27µl water
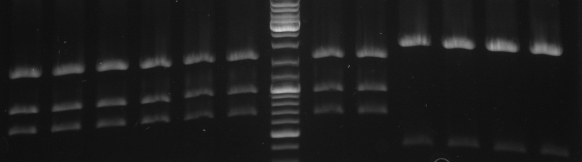
- Gel
- expected fragments: 292,601,2038bp
- lane0: DNA ladder mix
- lane1: 1.1
- lane2: 1.2
- lane3: 1.3
- lane4: 2.1
- lane5: 2.2
- lane6: 2.3
- lane7: 3.1
- lane8: 3.2
- lane9: 3.3
- lane10: 4.1
- lane11: 4.2
- lane12: 4.3
- mutagenesis pcr with 2.2 and 2.3 overnight to remove EcoRI restriction site
5µl Pfu buffer 2µl CmR_EcoRI_mut_fw 2µl CmR_EcoRI_mut_rv 1,5µl dNTPs 1µl Miniprep, diluted 1:10 1µl Pfu (not turbo!) 37.5µl water
PCR protocol 95°C 30s 95°C 30s | 55°C 45s | 16x 68°C 6.5min | 4°C for ever
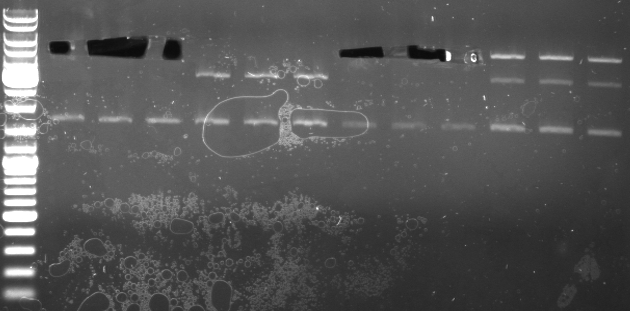
Phage cloning strategy two
- screening pcr with CmR_prefix_fw and CmR_suffix_rv primer (using Taq)
- Gel
- lane0: dna ladder
- lane1: colony 1+2
- lane2: colony 3+4
- lane3: colony 5+6
- lane4: colony 7+8
- lane5: colony 9+10
- lane6: colony 11+12
- lane7: colony 13+14
- expected sizes:
- pBlue with insert: 1668bp
- CmR cassette only: 852bp
- GFP cassette only: 919bp
- expected sizes:
- colonys 1,2,3,4,9,10 look good
Friday, 10/10/08
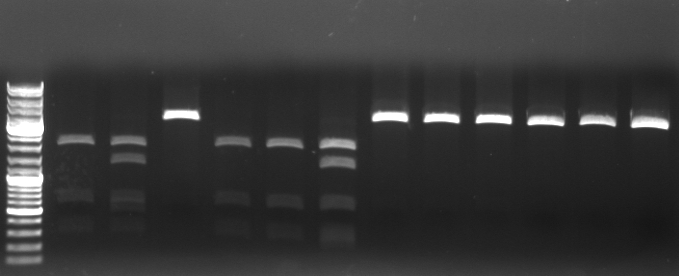
Cloning of CmR in standard plasmid
- proceeding of EcoRI mutagenesis pcr
- 3h DpnI digestion at 37°C
- transformation in TOP10
- digestion of the 12 minipreps (CmR in pSB1A2) with PstI/XbaI
- expected fragments: 2053bp, 878bp
- Gel
- lane0: ladder
- lane1-12: miniprep 1.1 - 4.3
--> miniprep 1.1, 2.1, 2.2 look good
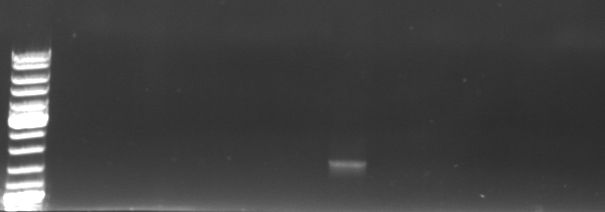
Phage cloning strategy two
- Screening PCR wirh oriT_fw and CmR_suffix_rv (using Taq)
- expected sized:
- pBlue/insert: 2150bp
- pBlue/GFP/CmR: ca. 1.3kb
- expected sized:
- Gel
- lane0: DNA ladder mix
- lane1-12: sample 1-12
- lane 7 looks good ~1.3kb
Saturday, 10/11/08
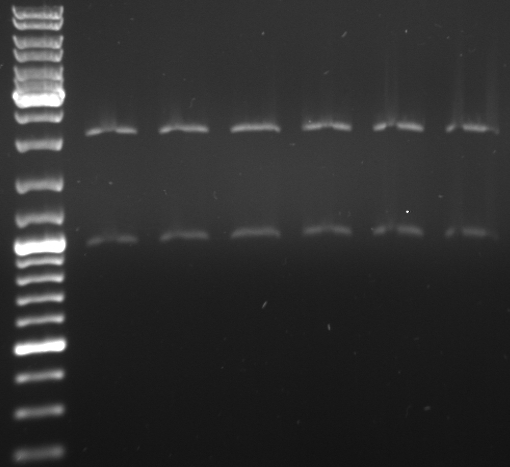
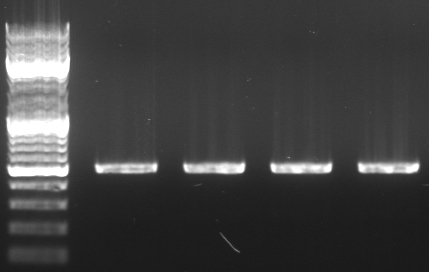
Cloning of oriT in standard plasmid
- Miniprep of oriT (from RP4)
- PCR with oriT prefix/suffix primer using Phusion
- expected size: ~500bp
- 4 pcr samples
- lane0: dna ladder mix
- lane1-4: oriT pcr probe 1-4
- PCR purification kit
- Digestion with PstI/EcoRI
- PCR purification kit
- Ligation in pSB1A2 20min at RT (after 40min heat inactivation at 65°C)
- Transformation
- transformation was done on Cm plates although there is no Cm resistance (failure!)
- -->make transformation again on sunday and plate out on Amp plates
Proceeding of cloning CmR in standard plasmid
- inoculation of Mutagensis PCR 2.2 (Cmr Std) (only one colony was grown)
- Mutagenesis PCR of 1.1, 1.2 and 2.2 (CmR Std) using Turbo Pfu
5µl Pfu buffer 2µl CmR_EcoRI_mut_fw 2µl CmR_EcoRI_mut_rv 1,5µl dNTPs 1µl Miniprep, diluted 1:10 1µl turbo Pfu 37.5µl water
PCR protocol 95°C 30s 95°C 30s | 55°C 45s | 16x 68°C 7min | 4°C for ever
Phage cloning strategy two
- inoculation of pBluescript+GFP+CmR 1-6,9,10 (nach fraktioniertem Ausstrich)
- redo Screening PCR wirh oriT_fw and CmR_suffix_rv (using Taq)
- no bands visible! (only primers)
Sunday, 10/12/08
Proceeding of cloning CmR in standard plasmid
- Miniprep of Mutagenesis PCR 2.2
- Digestion of Mutagenesis PCR Miniprep 2.2 with PstI, EcoRI
- -->digestion showed that the mutagenesis PCR did not work
- DpnI digestion of mutagenesesis PCR (2h at 37°C)
- transformation of mutagenesis PCR in TOP10
Proceeding of cloning oriT in standard plasmid
- redo transformation and plate out on Amp plates
Phage cloning strategy two
- Miniprep of pBlue+GFP+CmR 1-6,9,10
| << Week 9 | Overview | Week 11 >> |
|---|
 "
"