Team:USTC/Results
From 2008.igem.org
(Difference between revisions)
(→Correlation between GFP expression and cell density) |
m (→Correlation between GFP expression and cell density) |
||
| (15 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
|} | |} | ||
| - | {| style="color:#FFFFFF;background-color:#66B3FF;" cellpadding="1" cellspacing="2" border="3" bordercolor="#555" width=" | + | {| style="color:#FFFFFF;background-color:#66B3FF;" cellpadding="1" cellspacing="2" border="3" bordercolor="#555" width="870px" align="center" |
|- | |- | ||
!align="center"|[[Team:USTC|Home]] | !align="center"|[[Team:USTC|Home]] | ||
| Line 34: | Line 34: | ||
=== Correlation between GFP expression and cell density === | === Correlation between GFP expression and cell density === | ||
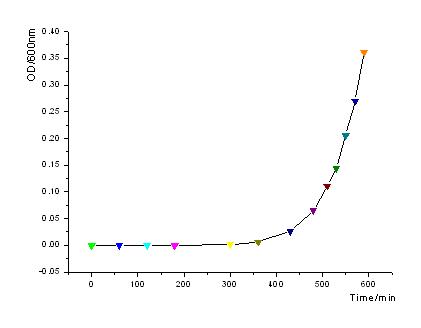
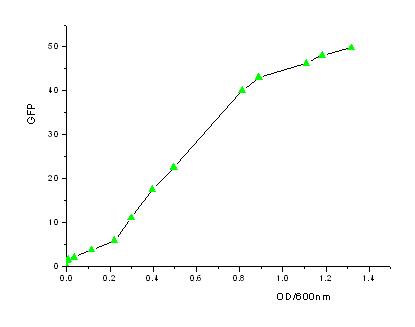
| - | + | In order to know how the expression level of signal molecules is related to cell density, we use GFP as the intermediate variable between the two variables. First, we determined the correlation of GFP expression according to the population of bacteria. Here, we have constructed the part with BioBricks R0040 and E0840. After ligating them together, we measured the amount of GFP expression and corresponding OD values and then worked out the correlation curve using Origin 7.5. | |
::[[Image:USTC_OD-time.jpg |450px|left|thumb|Fig1: OD600 increases exponently as to time.]] | ::[[Image:USTC_OD-time.jpg |450px|left|thumb|Fig1: OD600 increases exponently as to time.]] | ||
| Line 46: | Line 46: | ||
==Function Test == | ==Function Test == | ||
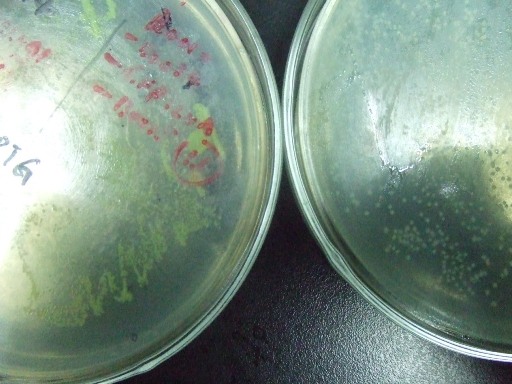
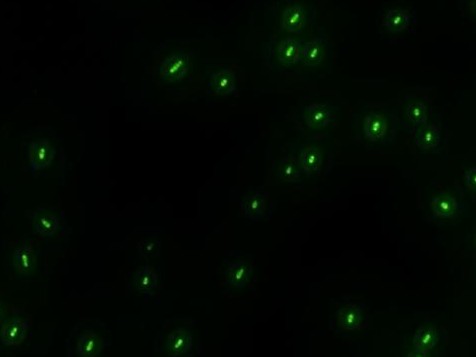
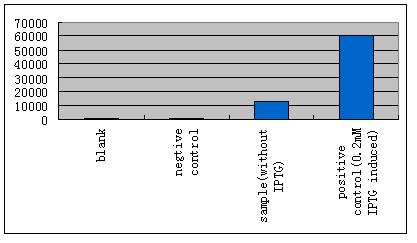
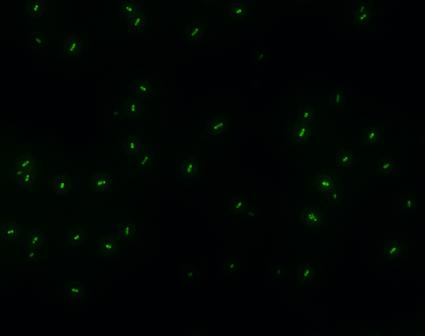
| - | *'''A'''fter GFP(LVA) | + | *'''A'''fter successfully cloning GFP(+LVA), we try to test its function.We ligate it with lac promoter R0010,a strong RBS B0034 and teminator B0015. We transform the plasmid with these parts into E.coli strain TOP10 and induce the expression of GFP with 0.1mM IPTG. |
| - | :::::[[Image:USTC-result1.jpg|left|300px|thumb| | + | :::::[[Image:USTC-result1.jpg|left|300px|thumb|Fig 3.Cells grow on the left plate generate GFP after IPTG induced,celles on the right plate is negtive control.]] |
| - | :::::::::::::::::::[[Image:USTC-result2.jpg|left|300px|thumb| | + | :::::::::::::::::::[[Image:USTC-result2.jpg|left|300px|thumb|Fig 4. Cells that generate GFP in fluorescence microscope]] |
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| - | *'''B'''ecause LacI is recruited here to regulate our system, IPTG | + | *'''B'''ecause LacI is recruited here to regulate our system, IPTG should be absent. As it is already known that the E.coli strain TOP10 expresses LacI constitutively at a basal level, we performed some experiments to test whether the influence can be small enough to be ignored. |
| - | [[Image:USTC-result3.jpg|left|350px|thumb| | + | |
| - | [[Image:USTC-result4.jpg|left|250px|thumb| | + | [[Image:USTC-result3.jpg|left|350px|thumb|Fig 5. This figure shows that though lacI gene exists in E.coli genome, GFP still is expressed without IPTG because of the high copy numbers of the plasmid. ]] |
| - | [[Image:USTC-result5.jpg|left|250px|thumb| | + | |
| + | [[Image:USTC-result4.jpg|left|250px|thumb|Fig 6.Cells express GFP with IPTG induced. ]] | ||
| + | |||
| + | [[Image:USTC-result5.jpg|left|250px|thumb|Fig 7.Though much weaker,Cells do express GFP under control of Lac promoter without IPTG. ]] | ||
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
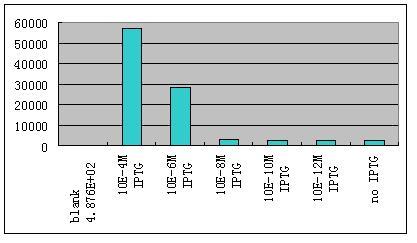
| - | * '''W'''e have constructed some | + | *'''W'''e then ligate this unit with the unit R0062-B0034-K082005-B0015 which gives weak constantly expression of a weak LacI mutant LacIM1.We use different concentration of IPTG to induce the expression of GFP.Results are shown as Fig 8. |
| + | ::::::::::[[Image:USTC-result6.jpg|left|350px|thumb|Fig 8]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | * '''W'''e have constructed some sequences with the purpose of testing whether the RNA polymerase can transcribe the target genes. For instance, we have built the following parts: | ||
:::# R0040-E0840; | :::# R0040-E0840; | ||
| Line 63: | Line 69: | ||
| - | :As expected, the plasmid containing the first three sequences can generate green fluerescence while the view field remains dark when the | + | :As expected, the plasmid containing each of the first three sequences can generate green fluerescence while the view field remains dark when the plasmid including the fourth sequence is present in bacteria. The results showed: |
| + | :::# ptetR (R0040) is a constitutively expressed promoter; | ||
| + | :::# At least the RNA polymerase can transcribe the sequence of C0070 and K082004; | ||
| + | :::# prhlR (R0071) cannot be transcribed by RNA polymerase alone. | ||
<br> | <br> | ||
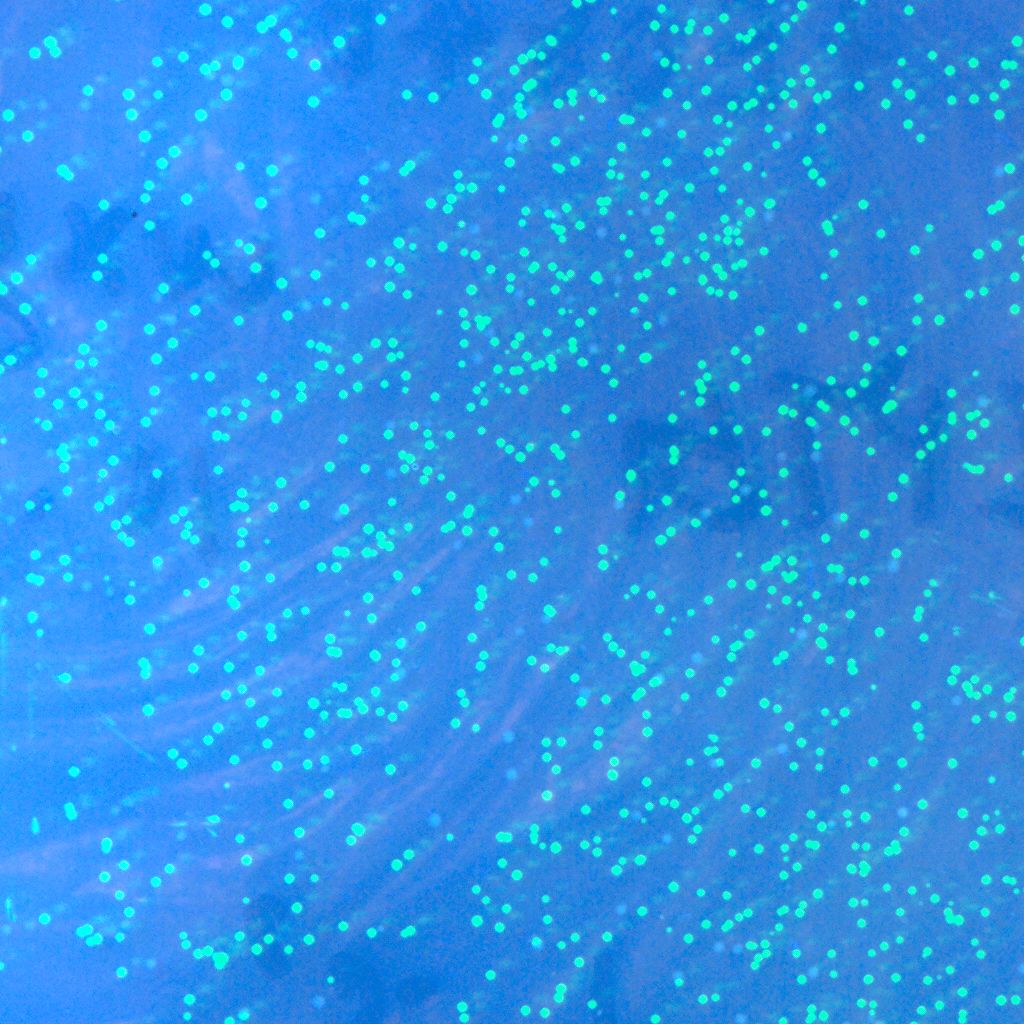
| - | * '''T'''he recombinase system is successfully constructed | + | * '''T'''he recombinase system is now successfully constructed, which can be shown by the two pictures below. The left one contains Cre+GFP, and the right one is lox71+GFP+lox66. They both express GFP as we expect. And the co-transformation of these two plasmids will be done in the next step. |
| - | :::::[[Image:USTC-gfp_1.jpg|left|300px|thumb|Fig | + | :::::[[Image:USTC-gfp_1.jpg|left|300px|thumb|Fig 9.Co-expression with Cre and GFP.(taken by Lu Xie)]] |
| - | :::::::::::::::::::[[Image:USTC-gfp2.jpg|left|300px|thumb|Fig | + | :::::::::::::::::::[[Image:USTC-gfp2.jpg|left|300px|thumb|Fig 10.Expression of GFP on the lox71+lox66 system.(taken by Lu Xie)]] |
Latest revision as of 04:00, 30 October 2008
| Home | The Team | The Project | Components | Results | Parts Submitted to the Registry | Notebook |
|---|
Correlation between GFP expression and cell density
In order to know how the expression level of signal molecules is related to cell density, we use GFP as the intermediate variable between the two variables. First, we determined the correlation of GFP expression according to the population of bacteria. Here, we have constructed the part with BioBricks R0040 and E0840. After ligating them together, we measured the amount of GFP expression and corresponding OD values and then worked out the correlation curve using Origin 7.5.
- From the figures above, we can draw the conclusion that the expression of GFP generally takes on a linear function of OD when the OD600 is between 0.2 to 0.9.
Function Test
- After successfully cloning GFP(+LVA), we try to test its function.We ligate it with lac promoter R0010,a strong RBS B0034 and teminator B0015. We transform the plasmid with these parts into E.coli strain TOP10 and induce the expression of GFP with 0.1mM IPTG.
- Because LacI is recruited here to regulate our system, IPTG should be absent. As it is already known that the E.coli strain TOP10 expresses LacI constitutively at a basal level, we performed some experiments to test whether the influence can be small enough to be ignored.
- We then ligate this unit with the unit R0062-B0034-K082005-B0015 which gives weak constantly expression of a weak LacI mutant LacIM1.We use different concentration of IPTG to induce the expression of GFP.Results are shown as Fig 8.
- We have constructed some sequences with the purpose of testing whether the RNA polymerase can transcribe the target genes. For instance, we have built the following parts:
- R0040-E0840;
- R0040-B0034-C0070-E0840;
- R0040-B0034-K082004-E0840;
- R0071-E0840.
- As expected, the plasmid containing each of the first three sequences can generate green fluerescence while the view field remains dark when the plasmid including the fourth sequence is present in bacteria. The results showed:
- ptetR (R0040) is a constitutively expressed promoter;
- At least the RNA polymerase can transcribe the sequence of C0070 and K082004;
- prhlR (R0071) cannot be transcribed by RNA polymerase alone.
- The recombinase system is now successfully constructed, which can be shown by the two pictures below. The left one contains Cre+GFP, and the right one is lox71+GFP+lox66. They both express GFP as we expect. And the co-transformation of these two plasmids will be done in the next step.
 "
"