Team:Harvard/Parts/LacI
From 2008.igem.org
(→Project Overview) |
(→LacI Inducible System) |
||
| (32 intermediate revisions not shown) | |||
| Line 55: | Line 55: | ||
{|align="justify" style="background-color:#FFFFFF;text-indent: 15pt;text-align:justify" cellpadding="50" width="90%" | {|align="justify" style="background-color:#FFFFFF;text-indent: 15pt;text-align:justify" cellpadding="50" width="90%" | ||
| | | | ||
| - | = | + | =LacI Inducible System= |
| - | + | In this system, the lac repressor (LacI) is controlled by a strong constitutive promoter, and is upstream of mtrB under the control of pLac, a LacI regulated promoter. In the default state, LacI is expressed, and inhibits transcription at pLac. This should allow us to control the expression of mtrB. In the default state, mtrB is not expressed. IPTG (an analog of allolactose) induces mtrB expression by binding to LacI, thereby preventing it from inhibiting transcription at pLac. | |
| + | |||
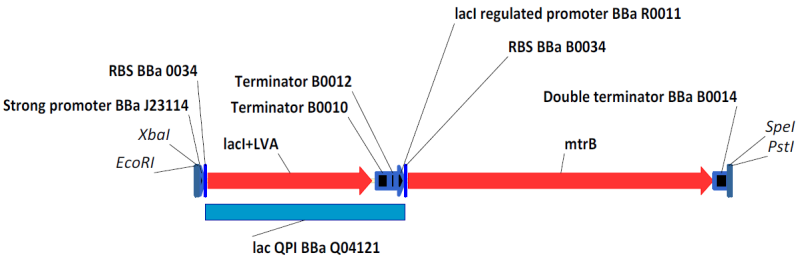
| + | <div style="text-indent:0pt;color:black">[[Image:BBa_K098984.png|thumb|650px|center|[http://partsregistry.org/Part:BBa_K098984 BBa_K098984] with BioBrick Prefix and Suffix, an example of the circuity we engineered. [http://partsregistry.org/Part:BBa_K098983 BBa_K098983] is similar, with a weaker promoter driving lacI expression.]]</div> | ||
| + | |||
| + | We didn't have enough time to finish thoroughly testing the above system, as the cloning of mtrB (toxic in ''E. coli'') was rather difficult. Our preliminary findings are quite interesting, though, so check out our [[Team:Harvard/Project| Project page]]. | ||
| + | |||
| + | ==Induction Test for LacI System with GFP== | ||
| + | In preparing to make the lacI system for mtrB, we tested the IPTG inducibility of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K098982 BBa_K098982] in ''E. coli''. In this system, the repressor is driven by a weak promoter. | ||
| + | ===Method=== | ||
| + | |||
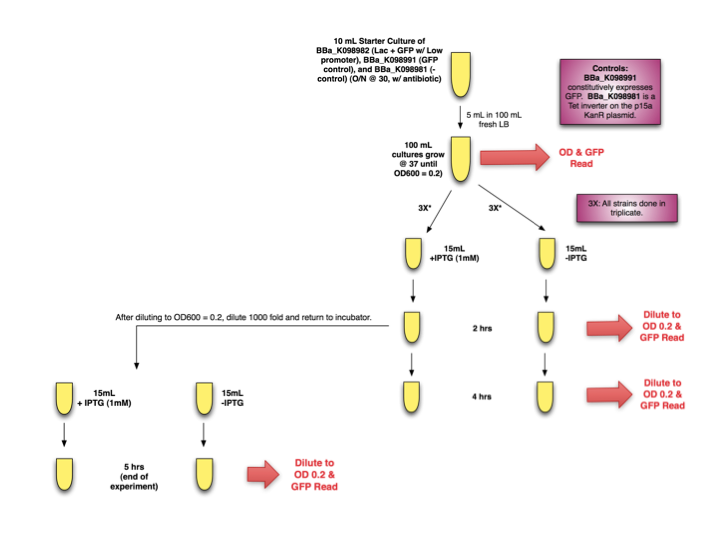
| + | Our test method is diagrammed below. | ||
| + | <div style="text-indent:0pt;color:black">[[Image:Lac.png|thumb|800px|center|]]</div><br> | ||
| + | |||
| + | ===Results=== | ||
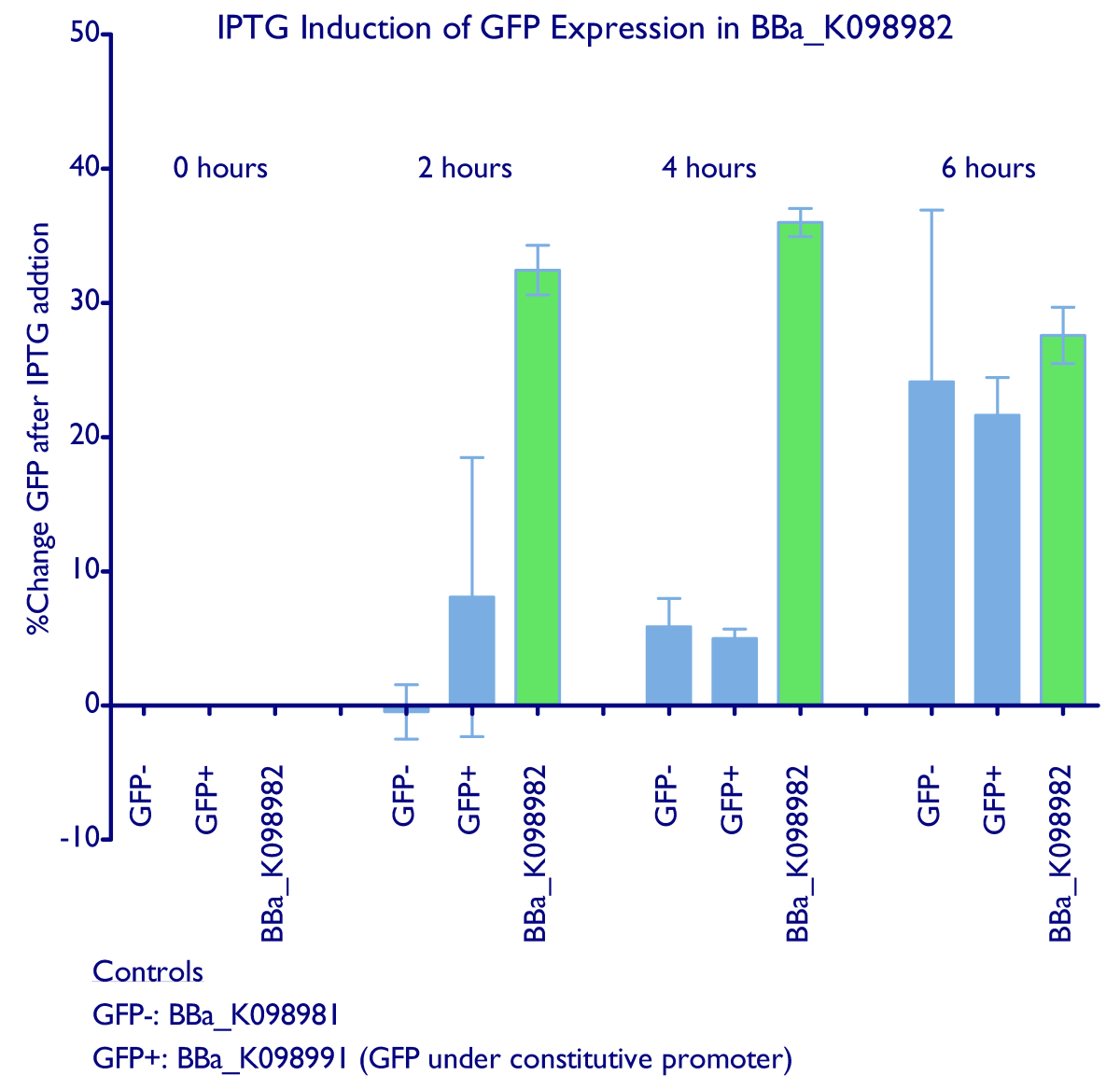
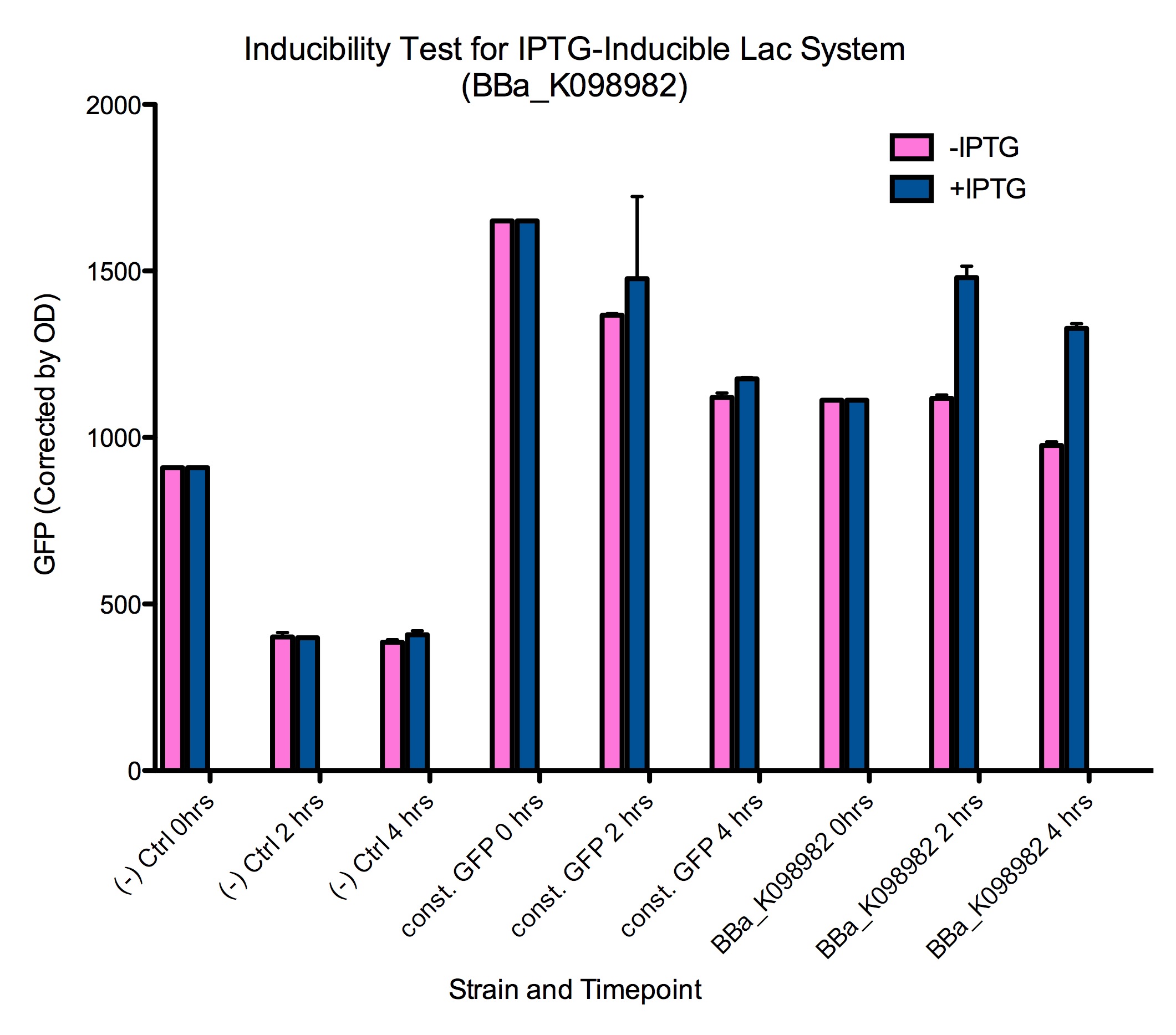
| + | Induction of GFP expression was observed at both 2 and 4 hours after adding IPTG. Levels of GFP expression in uninduced samples, however, remained relatively the same throughout the 4 hours. Meanwhile, IPTG induction was not observed in either the negative control ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K098981 BBa_K098981]) or the constitutive GFP control ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K098991 BBa_K098991]). Note that in the graph below, it appears that such an induction method is not reliable at 6 hours. | ||
| + | |||
| + | |||
| + | <div style="text-indent:0pt;color:black">[[Image:Baseline-corrected_of_lac_system.png|720px|thumb|center|IPTG successfully induces higher levels of GFP expression in cells containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K098982 BBa_K098982]<br>This data is also available in [https://static.igem.org/mediawiki/2008/f/fa/Baseline-corrected_of_lac_system.pdf PDF format].]]</div> | ||
| + | |||
| + | |||
| + | |||
| + | Additionally, it appears that even the uninduced state of such a system has significant expression of GFP. Tighter control may require higher levels of lacI expression. This leakiness may explain the difficulty we had in cloning mtrB (a gene toxic to ''E. coli''). | ||
| + | |||
| + | <div style="text-indent:0pt;color:black">[[Image:Lac.jpg|650px|thumb|center|IPTG induction raw data]]</div> | ||
| - | |||
| - | |||
| - | |||
|} | |} | ||
Latest revision as of 04:53, 30 October 2008
|
|
 "
"