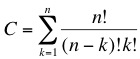Team:ETH Zurich/Project/Approach
From 2008.igem.org
(→Our Approach) |
(→Implementation) |
||
| (40 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<!-- PUT THE PAGE CONTENT AFTER THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT AFTER THIS LINE. THANKS :) --> | ||
| + | __TOC__ | ||
| + | == Random walk towards a minimal genome == | ||
| - | + | The concept we are trying to follow is based on a massively parallel random search. We have developed a top-down approach that will enable us to dramatically reduce the genome size of a living organism. As we described in the [[Team:ETH Zurich/Project/Background|previous section]] we are looking for a minimal subset of genes that are able to sustain life. The main problem when following this type of approach is the combinatorial possibilities that lead to a huge solution space, this corresponds to a classical NP-problem (non-polynomial) in computer science. The number of combinations of the original set of genes is given by: | |
| - | |||
| - | + | [[Image:NP.jpg|center]] | |
| - | + | This type of problem makes it infeasible to try all possible combinations sequentially and justifies a massively parallel approach as the one followed in this project. | |
| - | We | + | We propose the following algorithm as a method to find solutions to the presented problem: |
| - | + | Evolutionary algorithm: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | 1. Take the elements of current solution population (our populations in the chemostat). | |
| + | |||
| + | 2. Mutate them according to some evolutionary rules (pulse restriction enzymes and ligases). | ||
| + | |||
| + | 3. Test the fitting of each solution (grow in continuous culture under selective conditions). | ||
| + | |||
| + | 4. Bring only the fitter ones to next generation. | ||
| + | |||
| + | 5. Go to step 1 | ||
| + | |||
| + | |||
| + | The effect of this algorithm when implemented ''in vivo'' should be the following: | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:randomwalk.png|center]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | As can be seen in the previous figure, we start with our wild type genome and generate a large library of mutants by randomly deleting fragments of the chromosome as described in the [[Team:ETH Zurich/Wetlab/Genome Reduction|genome reduction]] section. | ||
| + | |||
| + | We then follow the new "paths" to test the fitness of our solutions and select for the ones that have successfully reduced the genome. These solutions are amplified by our continuous culture environment and represent the starting point for the subsequent round of mutations. | ||
| + | |||
| + | By iterating this process we should be able to follow a biased "random walk" towards the minimal genome and gradually reduce the genome of our target organism. | ||
| + | |||
| + | == Implementation == | ||
| + | |||
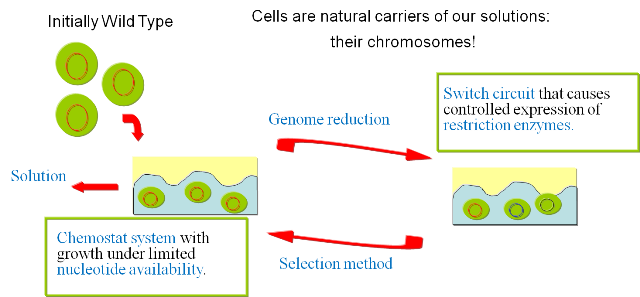
| + | In order to prove the viability of the concept, we have developed a series of models and biological experiments that are introduced in this section. For details on the results of the single steps we refer to the specific parts in the [[Team:ETH Zurich/Wetlab/Overview|wetlab]] and the [[Team:ETH Zurich/Modeling/Overview|modeling]] sections. The following figure gives an overview of the developed system and illustrates the final experimental setup. | ||
| + | |||
| + | |||
| + | [[Image:approach.png|center]] | ||
| + | |||
| + | |||
| + | We start by inoculating our continuous culture with a wild type strain with a knock out that makes it auxotroph for nucleotides. These nucleotides will be the limiting substrate in the chemostat and the concentration in our feed will be the parameter we can tune to set the selective pressure. See the [[Team:ETH Zurich/Wetlab/Chemostat Selection| chemostat selection]] section for a detailed description of the selection process. | ||
| + | |||
| + | The iteration of the following steps gradually reduces the size of the chromosome: | ||
| + | |||
| + | '''1) Excision of genomic sequences by restriction enzymes and ligases.''' We have developed a [[Team:ETH Zurich/Modeling/Switch Circuit|genetic circuit]] that allows us to generate a controlled pulse to control the expression of restriction enzymes and ligases. Our assumption is that by carefully selecting the timing of the pulse, we can cut fragments of the chromosome, that in a number of cases will be lost when the chromosome is religated. To test this principle in vivo we have developed a proof of concept that will show the potential viability of this method [[Team:ETH Zurich/Wetlab/Genome Reduction|(genome reduction)]]. | ||
| + | |||
| + | '''2) Growth in continuous culture under selective pressure.''' After the generation of the library of mutant we have to select for the strains that have successfully deleted part of the chromosome. Our [[Team:ETH Zurich/Modeling/Chemostat Selection|model]] predicts the effectiveness of the devised approach and supports our assumptions regarding the applicability of this method. | ||
| + | |||
| + | == Modeling framework == | ||
| + | |||
| + | A complete framework was developed to test our approach. We have implemented models of all steps of the presented method and integrated them in a framework that tries to capture the behavior of the system with the highest possible fidelity. | ||
| + | |||
| + | The different models include: | ||
| + | |||
| + | 1) The genome [[Team:ETH Zurich/Modeling/Genome Static Analysis|static analysis]]. Here we perform an exhaustive analysis of restriction enzymes based on a complete annotation of the e.coli K12 chromosome. We identify the different cutting characteristics and select for suitable candidates for subsequent simulations. | ||
| + | |||
| + | 2) A genome scale model of e.coli to perform [[Team:ETH Zurich/Modeling/Genome-Scale Model|flux balance]] analysis. With this part we simulate the effect of fragment deletions on viability and growth rate. | ||
| + | |||
| + | 3) A model of the [[Team:ETH Zurich/Modeling/Chemostat Selection|continuous culture]] that takes the output of the FBA to simulate the selection mechanism. | ||
| + | |||
| + | 4) A dynamic model of the [[Team:ETH Zurich/Modeling/Switch Circuit|pulse generator]] circuit based on the SimBiology toolbox. | ||
| + | |||
| + | == Wetlab proof of concept == | ||
| + | |||
| + | To assess the viability of our method in living organisms we have developed the following proofs of concept: | ||
| + | |||
| + | 1) [[Team:ETH_Zurich/Wetlab/Genome_Reduction|Genome Reduction]]. Here we study the possibility of ''in vivo'' restriction and religation of the chromosome. | ||
| + | |||
| + | 2) Our selection mechanism is based on the possibility to select organisms with different chromosomal sizes by limiting nucleotide availability. To test this assumption we have performed a series of growth experiments, the results are shown in the section dedicated to the [[Team:ETH_Zurich/Wetlab/Chemostat_Selection|chemostat selection]] section. | ||
| + | |||
| + | 3) The controlled expression of restriction enzyme and ligase is of fundamental importance for our project. We present the results of the [[Team:ETH_Zurich/Wetlab/Switch_Circuit|pulse generator]] implementation and the challenges we encountered. | ||
| - | |||
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Latest revision as of 03:34, 30 October 2008
Random walk towards a minimal genomeThe concept we are trying to follow is based on a massively parallel random search. We have developed a top-down approach that will enable us to dramatically reduce the genome size of a living organism. As we described in the previous section we are looking for a minimal subset of genes that are able to sustain life. The main problem when following this type of approach is the combinatorial possibilities that lead to a huge solution space, this corresponds to a classical NP-problem (non-polynomial) in computer science. The number of combinations of the original set of genes is given by:
We propose the following algorithm as a method to find solutions to the presented problem: Evolutionary algorithm: 1. Take the elements of current solution population (our populations in the chemostat). 2. Mutate them according to some evolutionary rules (pulse restriction enzymes and ligases). 3. Test the fitting of each solution (grow in continuous culture under selective conditions). 4. Bring only the fitter ones to next generation. 5. Go to step 1
We then follow the new "paths" to test the fitness of our solutions and select for the ones that have successfully reduced the genome. These solutions are amplified by our continuous culture environment and represent the starting point for the subsequent round of mutations. By iterating this process we should be able to follow a biased "random walk" towards the minimal genome and gradually reduce the genome of our target organism. ImplementationIn order to prove the viability of the concept, we have developed a series of models and biological experiments that are introduced in this section. For details on the results of the single steps we refer to the specific parts in the wetlab and the modeling sections. The following figure gives an overview of the developed system and illustrates the final experimental setup.
The iteration of the following steps gradually reduces the size of the chromosome: 1) Excision of genomic sequences by restriction enzymes and ligases. We have developed a genetic circuit that allows us to generate a controlled pulse to control the expression of restriction enzymes and ligases. Our assumption is that by carefully selecting the timing of the pulse, we can cut fragments of the chromosome, that in a number of cases will be lost when the chromosome is religated. To test this principle in vivo we have developed a proof of concept that will show the potential viability of this method (genome reduction). 2) Growth in continuous culture under selective pressure. After the generation of the library of mutant we have to select for the strains that have successfully deleted part of the chromosome. Our model predicts the effectiveness of the devised approach and supports our assumptions regarding the applicability of this method. Modeling frameworkA complete framework was developed to test our approach. We have implemented models of all steps of the presented method and integrated them in a framework that tries to capture the behavior of the system with the highest possible fidelity. The different models include: 1) The genome static analysis. Here we perform an exhaustive analysis of restriction enzymes based on a complete annotation of the e.coli K12 chromosome. We identify the different cutting characteristics and select for suitable candidates for subsequent simulations. 2) A genome scale model of e.coli to perform flux balance analysis. With this part we simulate the effect of fragment deletions on viability and growth rate. 3) A model of the continuous culture that takes the output of the FBA to simulate the selection mechanism. 4) A dynamic model of the pulse generator circuit based on the SimBiology toolbox. Wetlab proof of conceptTo assess the viability of our method in living organisms we have developed the following proofs of concept: 1) Genome Reduction. Here we study the possibility of in vivo restriction and religation of the chromosome. 2) Our selection mechanism is based on the possibility to select organisms with different chromosomal sizes by limiting nucleotide availability. To test this assumption we have performed a series of growth experiments, the results are shown in the section dedicated to the chemostat selection section. 3) The controlled expression of restriction enzyme and ligase is of fundamental importance for our project. We present the results of the pulse generator implementation and the challenges we encountered.
|
 "
"