Team:KULeuven/Model/Memory
From 2008.igem.org
(→Alternative) |
(→Parameters) |
||
| Line 69: | Line 69: | ||
|- | |- | ||
| K<sub>P2ogr</sub> | | K<sub>P2ogr</sub> | ||
| - | | 4.2156 | + | | 4.2156 |
| Used in two reactions for activator control at the transcription of P2ogr mRNA and CIIP22 mRNA | | Used in two reactions for activator control at the transcription of P2ogr mRNA and CIIP22 mRNA | ||
| [http://www.jbc.org/cgi/content/abstract/258/17/10536?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=P22+c2+repressor&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&volume=258&resourcetype=HWCIT link] | | [http://www.jbc.org/cgi/content/abstract/258/17/10536?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=P22+c2+repressor&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&volume=258&resourcetype=HWCIT link] | ||
|- | |- | ||
| K<sub>R0053_P22CII</sub> | | K<sub>R0053_P22CII</sub> | ||
| - | | 0.1099 | + | | 0.1099 |
| R0053 is the P22 cII promoter region | | R0053 is the P22 cII promoter region | ||
| | | | ||
Revision as of 14:07, 25 July 2008
Result:
To have a dropdown for your own team copy over all of the content below. It might be needed to adjust a thing or two. Comments have been added throughout the code what must be changed.
Inspirational websites:
- [http://javascript-array.com/scripts/simple_drop_down_menu/ Most basic dropdown menu, with no submenu's]
- [http://jquery.com/ The javascript library used to add effects and optimise the dropdown]
- [http://www.tyssendesign.com.au/articles/css/centering-a-dropdown-menu/ How to center a ul list effectively]
The dropdown has been created and developed by the KULeuven team.
<html>
<style type="text/css">
#content {z-index:4;}
#ddwrapper * {z-index:8 !important;}
div#ddwrapper {
margin:0;
padding:0;
height:28px;
width:945px; /* change to adjust imperfections in width */
}
div#ddnav {
margin:0 auto; /* needed to center the dropdown */
padding:0;
top:5px;
/* width: 965px */
visibility:hidden; /* dropdown is hidden until properly initalised by javascript */
}
div#ddtoggle {
margin:0;
position:fixed;
right:2px;
top:15px;
height:10px;
width:10px;
z-index:100;
}
#ddnav ul {
display:table-row; /* works only for firefox, later adjusted by javascript for IE */
margin:0 auto;
padding:0;
}
#ddnav ul li {
display:table-cell; /* works only for firefox, later adjusted by javascript for IE */
list-style:none;
margin:0;
padding:0 !important;
border-right:1px solid #FFF; /* creates illusion of spacing between tabs */
}
#ddnav ul li:last-child{border-right:none;}
#ddnav a{
display:block;
margin:0;
padding:4px 14px; /* play with dimensions for size of tabs */
background-color:#075A90; /* background color for tabs */
color:#FFF !important; /* font color for text in tabs */
text-align:center; /* aligning for text in tabs */
text-decoration:none !important;
font:bold 10pt Trebuchet MS; /* font properties for text in tabs */
outline:0;
}
#ddnav ul li a:hover {background-color:#99CCFF;}/* background color for tabs on mouseover */
#ddnav li a:active {outline:none;} /* remove standard dotted border for links when clicked (IE) */
#ddnav li a:focus {-moz-outline-style:none;} /* remove standard dotted border for links when clicked (FF) */
#ddnav div {
display:none;
position:absolute;
width:9em;
background-color:#000; /* bug solution, do not change ! */
border:1px solid #5970B2; /* border color for dropdown menus */
opacity:0.9; /* transparancy of the dropdown menus (FF) */
filter:alpha(opacity=90); /* transparancy of the dropdown menus (IE) */
}
#ddnav div a {
display:block;
padding:5px 10px; /* play with dimensions of block element in dropdown menus */
position:relative;
font:normal 8pt arial; /* font properties for text in dropdown menus */
text-align:left; /* aligning of text in dropdown menus */
cursor:pointer;
}
#ddnav div a:hover, #ddnav span a:hover {color:#000 !important;} /* text color on mouseover */
#ddnav span div {
position:relative;
border:none;
border-bottom:2px groove #5970B2; /* separator for submenus, groove does not work in FF */
opacity:1.0; /* avoid stacking transparancy for submenus (FF) */
filter:alpha(opacity=100); /* avoid stacking transparancy for submenus (IE) */
}
/* may want to upload the following pictures to a new location */
.expand {background: url('https://static.igem.org/mediawiki/2008/e/ef/Icon-expand.png') no-repeat 95% 50%;}
.collapse {background: url('https://static.igem.org/mediawiki/2008/c/cd/Icon-collapse.png') no-repeat 95% 50%;}
.docked {background: #99ccff url("https://static.igem.org/mediawiki/2008/6/62/Ddnavundock.png") no-repeat 50% 50%;}
.undocked {background: #99ccff url("https://static.igem.org/mediawiki/2008/e/e4/Ddnavdock.png") no-repeat 50% 50%;}
</style>
<!-- IMPORTANT: save following script under a personalized webspace or download the library at www.jquery.com -->
<script type="text/javascript" src="http://student.kuleuven.be/~s0173901/wiki/js/jquery.js"></script>
<script type="text/javascript">
function ddnav() {
$('#ddnav ul li').hover(
function () {
$(this).find('div:first').css('display','block');},
function () {
$(this).find('div:first').css('display','none');}
);
}
function ddnavsub() {
$('#ddnav span > a').toggle(
function () {
$(this).removeClass("#ddnav expand").addClass("#ddnav collapse");
$(this).parent().find('div:first').slideDown('fast');
$(this).hover(
function (){$(this).css('background-color','#99AAFF');},
function (){$(this).css('background-color','#99AAFF');});},
function () {
$(this).removeClass("#ddnav collapse").addClass("#ddnav expand");
$(this).parent().find('div:first').css('display','none');
$(this).hover(
function (){$(this).css('background-color','#99CCFF');},
function (){$(this).css('background-color','#075A90');});}
).addClass("#ddnav expand");
}
/* If you wish to omit the docking feature, remove following function ddtoggle() */
function ddtoggle() {
$('#ddtoggle').toggle(
function () {
$(this).removeClass('undocked').addClass('docked');
$('#ddnav').css('position','fixed');},
function () {
$(this).removeClass('docked').addClass('undocked');
$('#ddnav').css('position','static');}
);
}
function ddalign() {
var _windowWidth = $(window).width();
var _leftOffset = (_windowWidth - 965)/2;
$('div#ddnav').css('left',_leftOffset);
}
function ddmsie() {
$('#ddnav a').hover(
function () {$(this).css('background-color','#99ccff');},
function () {$(this).css('background-color','#075a90');}
);
/* toggle doesn't work yet */
$('#ddtoggle').css('display','none');
$('#ddnav ul').css('display','inline-block');
$('#ddnav ul li').css('display','inline');
$('#ddnav ul li').css('position','relative');
$('#ddnav ul li>a').css('display','inline-block');
$('#ddnav ul li>a').css('margin-right','-4px');
$('#ddnav div').css('left','0');
$('#ddnav div').css('top','100%');
$('#ddnav span div').css('top','0');
}
function ddmozilla() {
ddtoggle();
$(window).bind('resize', function() {ddalign();});
}
$(function () {
ddnav();
ddnavsub();
if(jQuery.browser.msie) ddmsie();
if(jQuery.browser.mozilla) ddmozilla();
$('#ddnav').css('visibility','visible');
});
</script>
<!-- If you wish to omit the docking feature omit following line (div with id ddtoggle) -->
<div id="ddtoggle" class="undocked"></div>
<div id="ddwrapper">
<!-- Here the actual links are defined, simply replace with your own links in the appropriate sections -->
<div id="ddnav" align="center">
<ul>
<li>
<a href="https://2008.igem.org/Team:KULeuven">Home</a>
</li>
<li>
<a>The Team</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Team/LabsandGroups">Research Labs and Groups</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Students">Students</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Instructors">Instructors</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Advisors">Advisors</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Pictures">Pictures</a>
</div>
</li>
<li>
<a>The Project</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Project">Summary</a>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Project/Input">Input</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Evaluation">End Evaluation</a>
<a href="https://2008.igem.org/Team:KULeuven/Literature">Literature</a>
<a href="https://2008.igem.org/Team:KULeuven/Brainstorm">Brainstorm</a>
</div>
</li>
<li>
<a>Ethics</a>
<div>
</div>
</li>
<li>
<a>Submitted Parts</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Parts">Listing</a>
<a href="http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=KULeuven">Sandbox</a>
</div>
</li>
<li>
<a>Modeling</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Model/Overview">Overview</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/KineticConstants">Kinetic Constants</a>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Model/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Model/FullModel">Full Model</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Sensitivity">Sensitivity Analysis</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/MultiCell">Multi-cell Model</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Diffusion">Diffusion</a>
</div>
</li>
<li>
<a>Data Analysis</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/Overview">Overview</a>
<span>
<a>New Parts</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/GFP">GFP (LVA-tag)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/T7">T7 (UmuD-tag)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Antisense">Antisense LuxI</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/ccdB">Celldeath (ccdB)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/HybridProm">Hybrid Promotor</a>
</div>
</span>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/Input">Input</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Data/FullModel">Full Model</a>
</div>
</li>
<li>
<a>Software</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Software/MultiCell">Multi-cell Toolbox</a>
<a href="https://2008.igem.org/Team:KULeuven/Software/Simbiology2LaTeX">Simbiology2LaTeX Toolbox</a>
</div>
</li>
<li>
<a>Notebook</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Meeting_Calendar">Calendar</a>
<a href="https://2008.igem.org/Team:KULeuven/SummerHolidays">Summer Holidays</a>
<span>
<a>Reports</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Meeting Reports">Daily</a>
<a href="https://2008.igem.org/Team:KULeuven/Weekly Meetings">Weekly</a>
</div>
</span>
<span>
<a>Lab Data</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Freezer">Freezer</a>
<a href="https://2008.igem.org/Team:KULeuven/Primers">Primers</a>
<a href="https://2008.igem.org/Team:KULeuven/Ligation">Ligation</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Tools">Tools</a>
<a href="https://2008.igem.org/Team:KULeuven/Press">Press</a>
<a href="https://2008.igem.org/Team:KULeuven/Guestbook">Guestbook</a>
</div>
</li>
</ul>
</div>
</div>
</html>
Contents |
Memory
Position in the system
Describing the system
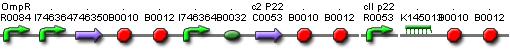
This system must activate the cell death system after one light pulse. As long as there is no light, there is no P2ogr, no CIIP22 and a lot of antimRNA_LuxI. The antimRNA_LuxI blocks the cell death system. When light is turned on OmpF increases. This causes P2ogr and CIIP22 to increase and antimRNA_LuxI to decrease. This activates the system. When light is turned off, the P2ogr concentration is large enough to maintain itself. This way antimRNA doesn't increase. The system stays activated.
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation Rates | |||
| dP2ogr | 0.002265 s-1 | ||
| dRNA_P2ogr | 0.002265 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dP22CII | 0.002311 s-1 | ||
| dRNA_P22CII | 0,0022651 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dantimRNA_luxI | 0.0045303 s-1 | ||
| Transcription Rates | |||
| kP2ogr | 0.0125 s-1 | ||
| kP22CII | 0.0125 s-1 | ||
| kAntimRNA_LuxI | 0.0094 s-1 | ||
| Dissociation Constants | |||
| KP2ogr | 4.2156 | Used in two reactions for activator control at the transcription of P2ogr mRNA and CIIP22 mRNA | [http://www.jbc.org/cgi/content/abstract/258/17/10536?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=P22+c2+repressor&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&volume=258&resourcetype=HWCIT link] |
| KR0053_P22CII | 0.1099 | R0053 is the P22 cII promoter region | |
| Hill Cooperativity | |||
| n | 2 | Used for all reactions throughout the memory submodel using Hill kinetics | |
Models
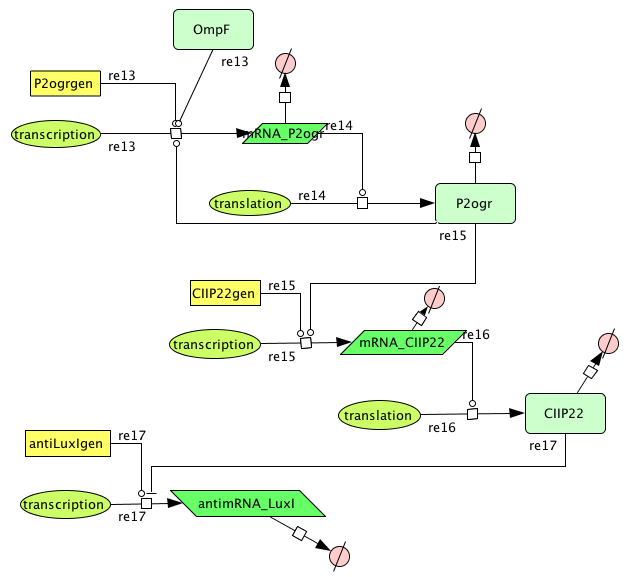
CellDesigner
- Figure: CellDesigner system representation
Matlab
Problem
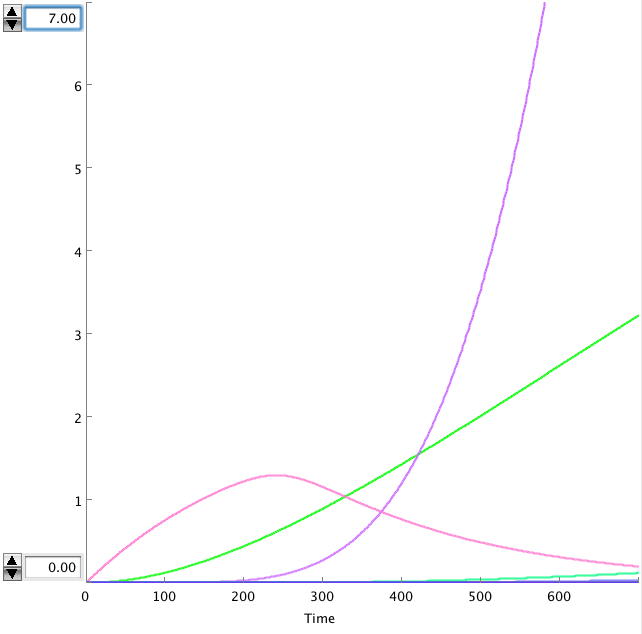
The OmpF promoter is not ideal. When there is no light the transcription rate is still 0.00005. This means that P2ogr will slowly build up, activating the system. In this case the memory is in 0-state when the stationary state isn't reached yet. The 1-state is the stationary state. So the system automatically ends up in state 1 after some time (300s). This can be seen in the figure below.
- Figure: Celldesigner simulation of the memory system. CIIP22(purple), P2ogr(green), AntimRNA(pink).
The system can only stay in 0-state for 300 sec. This makes it completely useless.
Alternative
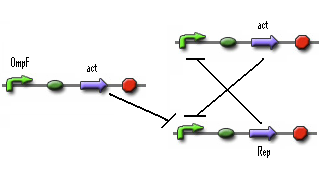
In the previous system the 0-state isn't actively maintained. It's just 'not stationary state'. So we need to search mathematical system that has 2 stationary states. A possible solution is given below.
- Figure: Part representation of alternative system
When this system starts Rep build up because Rep represses the Act promoter better then Act represses the Rep promoter. The Rep concentration stays high and the Act stays low. This is the 0-state. When there is light, the OmpF promoter is activated and the Act concentration is increased. This represses Rep promoter. The Rep concentration decreases and the Act promoter is activated. The Act concentration keeps increasing. When the light pulse ends the Act concentration is high enough to repress the Rep promoter, Act concentration stays high and Rep concentration stays low. This is the 1-state.
Using typical value this system can be tested in CellDesigner.
- Figure: Celldesigner system representation
These typical parameters are:
| Name | Value | ||
|---|---|---|---|
| Degradation Rates | |||
| dact | 0.0011552 s-1 | ||
| dRNA_act | 0.0023105 s-1 | ||
| drep | 0.0011552 s-1 | ||
| dRNA_rep | 0.0023105 s-1 | ||
| Transcription Rates | |||
| kact | 0.006 s-1 | ||
| krep | 0.0125 s-1 | ||
| Translation rate | |||
| kact | 0,166666 [M] | ||
| krep | 0,166666 [M] | ||
| Dissociation Constants | |||
| Kact-rep | 4.2156 [M] | ||
| Krep-act | 4.2156 [M] | ||
| Hill Cooperativity | |||
| n | 2 | ||
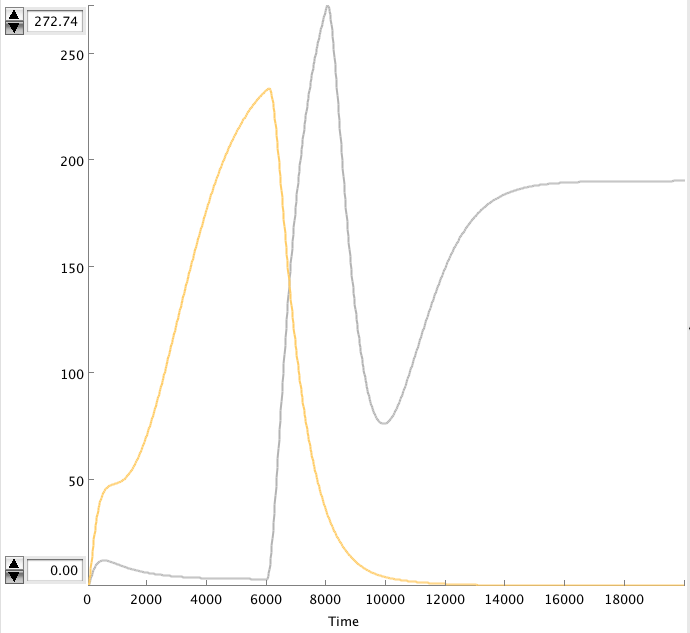
CellDesigner gives the following simulation when OmpF transcription rate changes from 0.0001 to 0.01 at t=6000 sec for 2000 sec.
- Figure: Celldesigner simulation of the alterative system. Act(grey), Rep(yellow)
The OmpF peak causes the Act concentration to rise and the Rep to decrease. The high Act concentration keeps the Rep concentration low. This causes the Act concentration to stay high.
 "
"