Team:Alberta NINT/Project
From 2008.igem.org
(→Terminators) |
m (→Project Details) |
||
| Line 91: | Line 91: | ||
<br><br> | <br><br> | ||
| - | [[image:NINT_Draw_ProjectDetails_HisTermDisrupted.jpg|thumb|400 px| | + | [[image:NINT_Draw_ProjectDetails_HisTermDisrupted.jpg|thumb|400 px|Anti-sense disruption of terminator]] |
=== Anti-sense disruption of terminator/attenuators === | === Anti-sense disruption of terminator/attenuators === | ||
The formation of terminator/attenuator (TA) stem-loop-stem hairpin structures can be disrupted using a second anti-sense) RNA input sequence. If the hybrid double-stranded RNA sequence formed by the RNA transcript and the anti-sense RNA input has greater stability (lower free energy) than the original haiprin TA structure, its formation will be preferred over the TA, permitting RNA transcription to proceed past the TA sequence. In the image (right), the input anti-sense sequence includes the entire 5' portion of the stem and the first three nucleotides in the loop. This is sufficient to confer an approximate 6 Kcal/mol stability advantage over the original TA secondary structure. This should cause this structure to be thermodynamically highly favored. | The formation of terminator/attenuator (TA) stem-loop-stem hairpin structures can be disrupted using a second anti-sense) RNA input sequence. If the hybrid double-stranded RNA sequence formed by the RNA transcript and the anti-sense RNA input has greater stability (lower free energy) than the original haiprin TA structure, its formation will be preferred over the TA, permitting RNA transcription to proceed past the TA sequence. In the image (right), the input anti-sense sequence includes the entire 5' portion of the stem and the first three nucleotides in the loop. This is sufficient to confer an approximate 6 Kcal/mol stability advantage over the original TA secondary structure. This should cause this structure to be thermodynamically highly favored. | ||
<br><br> | <br><br> | ||
| + | |||
| + | [[image:NINT_Draw_ProjectDetails_TermAntiTerm.jpg|thumb|400 px|Terminators and Anti-terminators]] | ||
| + | === Terminators and anti-terminators === | ||
| + | TA sequences can be preceded by competing sequences which preferentially form stem-loop-stem structures that are not followed immediately by U-rich terminating sequences. Such structures are called anti-terminators. If the stability of the anti-terminator structure is greater than that of the following terminator structure, it will be preferentially formed and prevent attenuation of the RNA transcript. In the image (right) a terminator (T) structure is out-competed by an anti-terminator structure (AT). The colors overlapping the sequence correlate to the colors in the cartoon of the structures. | ||
| + | <br><br> | ||
More Coming Soon! | More Coming Soon! | ||
Revision as of 07:55, 4 August 2008
| Home | The Team | The Project | Lab Protocols | Bits and Pieces | Modeling | Notebook |
|---|
Contents |
Manipulating RNA attenuator sequences to generate genetic logic circuits in E. coli...
Terminator/Attenuator anti-sense Logic (TAasL)
The integrated circuit was a major milestone of modern technology that facilitated the entry of society into the information age. Electrical circuits that once occupied an entire room are now able to be placed on a single micro-chip the size of a postage stamp. The implementation of Boolean logic (AND, OR, NOT, etc.) using integrated electronic circuits forms the basis for modern day computers.
When compared to electronics, similar levels of integration have yet to be realized in biological systems. While electronic devices are inherently connectible due to their common currency (flow of electrons), biological circuits lack similar connectivity. This poses a significant challenge to the development of truly programmable biosystems. Solving this challenge should open the door for development of biological devices just as integrated circuits lead to an explosion of electronic devices.
The Logi-coli[i] project hopes to overcome this challenge by constructing connectible, extensible biological circuits based on antisense RNA regulation of transcriptional attenuation. Transcriptional attenuation is a common biological mechanism for controlling transcriptional activity. As RNA strands are produced by RNA polymerase transcription of a DNA template, they often form secondary “stem-loop” structures due to base-pair complementarity of inverted repeat sequences. Inverted repeat sequences are short sequences in the transcript that are repeated at a later position in reverse order. These repeat sequences cause the RNA strand to fold back on itself due to base-pairing between complementary A-U and C-G nucleotide pairs in the different repeat regions. A terminator is formed at the end of an RNA transcript when such a simple stem-loop is followed by a “UUUUUUU” sequence that causes the severing of the nascent RNA strand and the dislocation of the RNA polymerase from the DNA template strand. An attenuator is a terminator structure found near the start of a transcript and has the ability to prematurely truncate the transcript.
We hypothesize that transcriptional attenuation can be modulated by the action of an anti-sense RNA strand disrupting the normal base-pairing in the attenuator stem-loop structure. As a result of this disruption, the RNA polymerase can continue transcription past the attenuator sequence. Using software called RNAstructure4.5, our team hopes to be able to rationally design functional attenuator and corresponding input anti-sense RNA sequences. In this way, we hope to provide a means for connecting series of circuits by using the output RNA strand of one circuit to regulate the transcription of a second circuit. We hope to combine simple attenuators into more complex structures that act as biological equivalents to electronic Boolean logic gates (AND, OR, XOR, etc.)
Thus, our scheme will be connectible because the output from one component (an RNA transcript) will be able to serve as the input to another component (the next TA gate). In addition, the scheme should provide for extensibility as it should be possible to rationally design a fairly large number of TA gates and anti-sense input/output signals with minimum cross-talk.
In regards to the final output, a LacZ coding region will be expressed to provide evidence of the functionality of the logic circuits. We hope to connect these basic logic gates into even more complex circuits such as half-adders or full-adders. This important step in the design of complex logic in biological circuits has eluded researchers for a number of years, but we believe our approach to have great potential for success. Potential applications for logical circuitry in organisms include almost any sensory, patterning or information processing scenario. Simple sensory circuits could potentially be combined to signal particular mixtures of molecules. Medical microbots could process complex chemical signals inside patient tissues and autonomously make therapeutic decisions based on their programming. Synthetic tissue could make patterning decisions to form biological shapes never seen in nature. Trying to describe the potential applications of programming behavior into living organisms is as difficult as describing the computer, cell phone or HDTV to a society which has just invented the lightbulb. Although we may be able to scratch the surface of the potential, it is certain that many unforeseen inventions will be generated in the future.
Project Details
Terminators
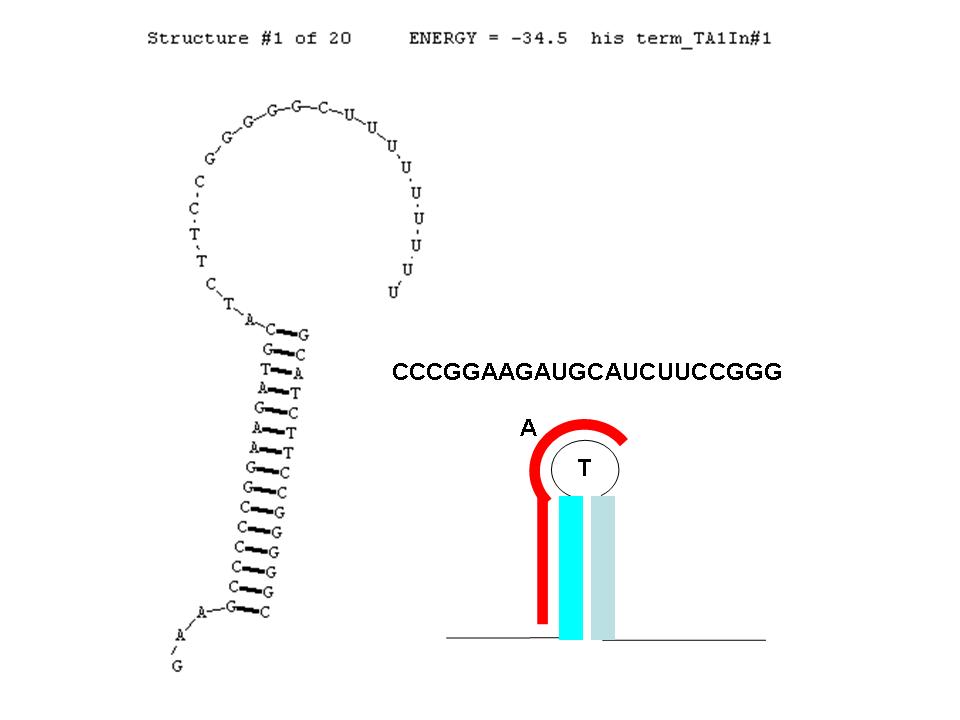
A classic terminator found at the 3' end of an RNA transcript consists of a "stem" structure of length ~10 to 30 nucleotides followed by a "loop" of 4-6 nucleotides, then several complementary nucleotides forming the corresponding 3' portion of the stem. Finally a weakly-hybridizing sequence of about 9 nucleotides (mainly U's or A's) forms the portion that permits the nascent transcript to dissociate from the template DNA strand. The termination efficiency (TE) of this structure is determined by many complex factors, including its thermodynamic stability. For this structure (see right) this free energy is calculated by RNAStructure as -28.5 Kcal/mole. The inset image is a cartoon representation of the terminator. Similar images will be used throughout this description.
Anti-sense disruption of terminator/attenuators
The formation of terminator/attenuator (TA) stem-loop-stem hairpin structures can be disrupted using a second anti-sense) RNA input sequence. If the hybrid double-stranded RNA sequence formed by the RNA transcript and the anti-sense RNA input has greater stability (lower free energy) than the original haiprin TA structure, its formation will be preferred over the TA, permitting RNA transcription to proceed past the TA sequence. In the image (right), the input anti-sense sequence includes the entire 5' portion of the stem and the first three nucleotides in the loop. This is sufficient to confer an approximate 6 Kcal/mol stability advantage over the original TA secondary structure. This should cause this structure to be thermodynamically highly favored.
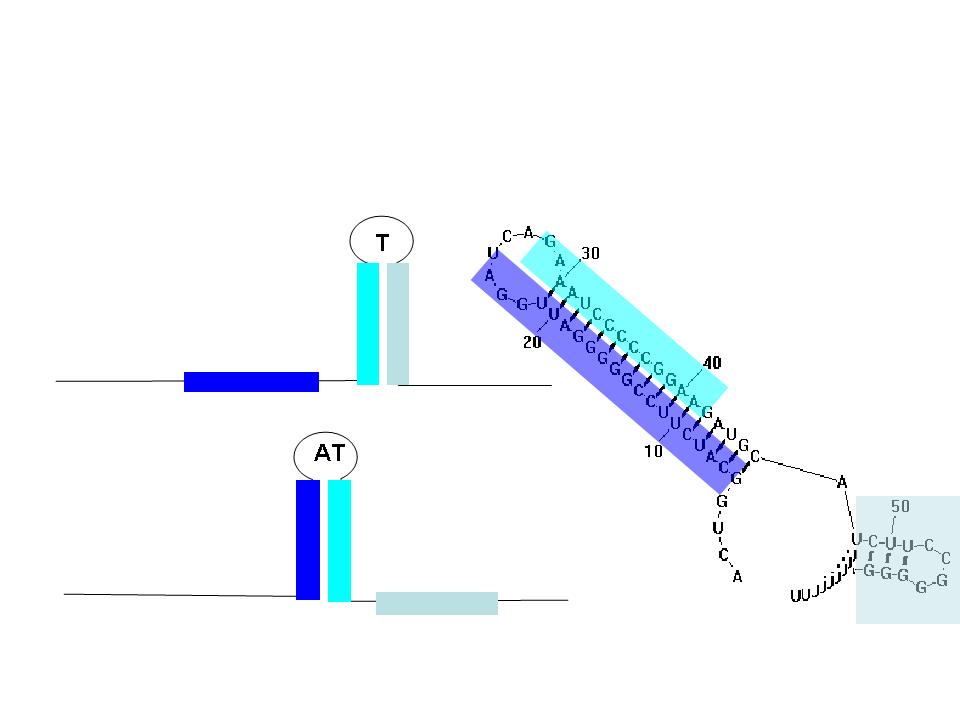
Terminators and anti-terminators
TA sequences can be preceded by competing sequences which preferentially form stem-loop-stem structures that are not followed immediately by U-rich terminating sequences. Such structures are called anti-terminators. If the stability of the anti-terminator structure is greater than that of the following terminator structure, it will be preferentially formed and prevent attenuation of the RNA transcript. In the image (right) a terminator (T) structure is out-competed by an anti-terminator structure (AT). The colors overlapping the sequence correlate to the colors in the cartoon of the structures.
More Coming Soon!
 "
"