Team:Wisconsin/Project
From 2008.igem.org
| Line 21: | Line 21: | ||
|Fuel consumption has come to the forefront as an important political and biological issue that has lead to innovative pursuits of renewable fuel as well as the controversial exploitation of natural resources. Currently ethanol is the commercial biofuel of choice; however, currently production and distillation of ethanol is inefficient. With this problem in mind, iGEM Wisconsin has looked for alternative ways to make not only ethanol, but other biofuels through synthetic biology. We've designed the following two projects using ''E. coli'' to produce biofuels in innovative ways: | |Fuel consumption has come to the forefront as an important political and biological issue that has lead to innovative pursuits of renewable fuel as well as the controversial exploitation of natural resources. Currently ethanol is the commercial biofuel of choice; however, currently production and distillation of ethanol is inefficient. With this problem in mind, iGEM Wisconsin has looked for alternative ways to make not only ethanol, but other biofuels through synthetic biology. We've designed the following two projects using ''E. coli'' to produce biofuels in innovative ways: | ||
| - | [[Image: | + | [[Image:Flowchart.png|800px]] |
One project focuses on using ''E. coli'' to produce sorbitol, a sugar alcohol, in large quantities for eventual commercial scale catalytic conversion to hydrocarbons. Along with producing sorbitol, we've modeled alterations in ''E. coli'' to make sorbitol production from a glycerol carbon source possible. Our aim is to modify a cell that will utilize glycerol, a byproduct from biodiesel production, and effectively create sorbitol. | One project focuses on using ''E. coli'' to produce sorbitol, a sugar alcohol, in large quantities for eventual commercial scale catalytic conversion to hydrocarbons. Along with producing sorbitol, we've modeled alterations in ''E. coli'' to make sorbitol production from a glycerol carbon source possible. Our aim is to modify a cell that will utilize glycerol, a byproduct from biodiesel production, and effectively create sorbitol. | ||
| Line 38: | Line 38: | ||
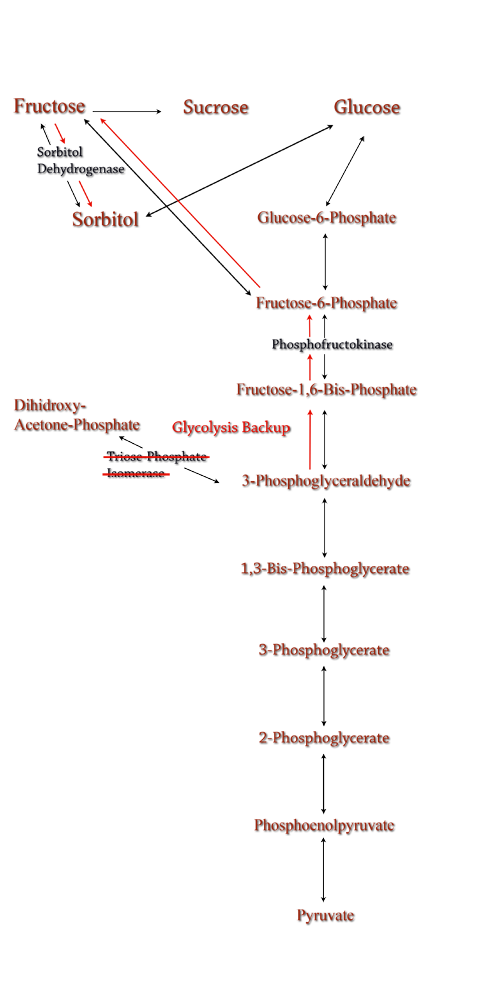
[[Image:TPI-Knockout.png|250px|thumb|TPI Knockout]] | [[Image:TPI-Knockout.png|250px|thumb|TPI Knockout]] | ||
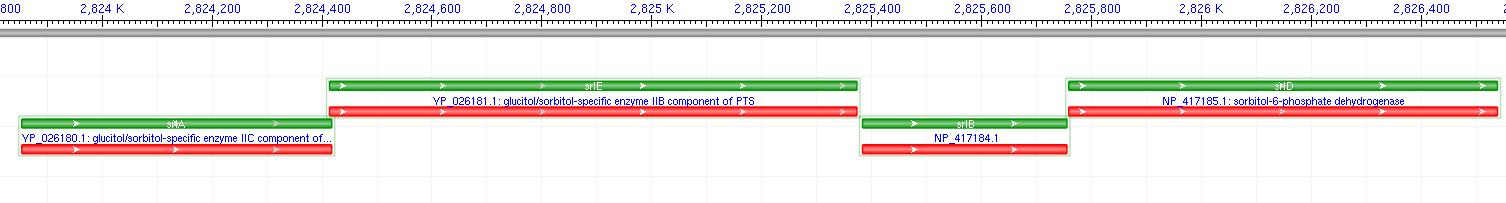
[[Image:SRLOp.JPG|800px|thumb|SRL Operon]] | [[Image:SRLOp.JPG|800px|thumb|SRL Operon]] | ||
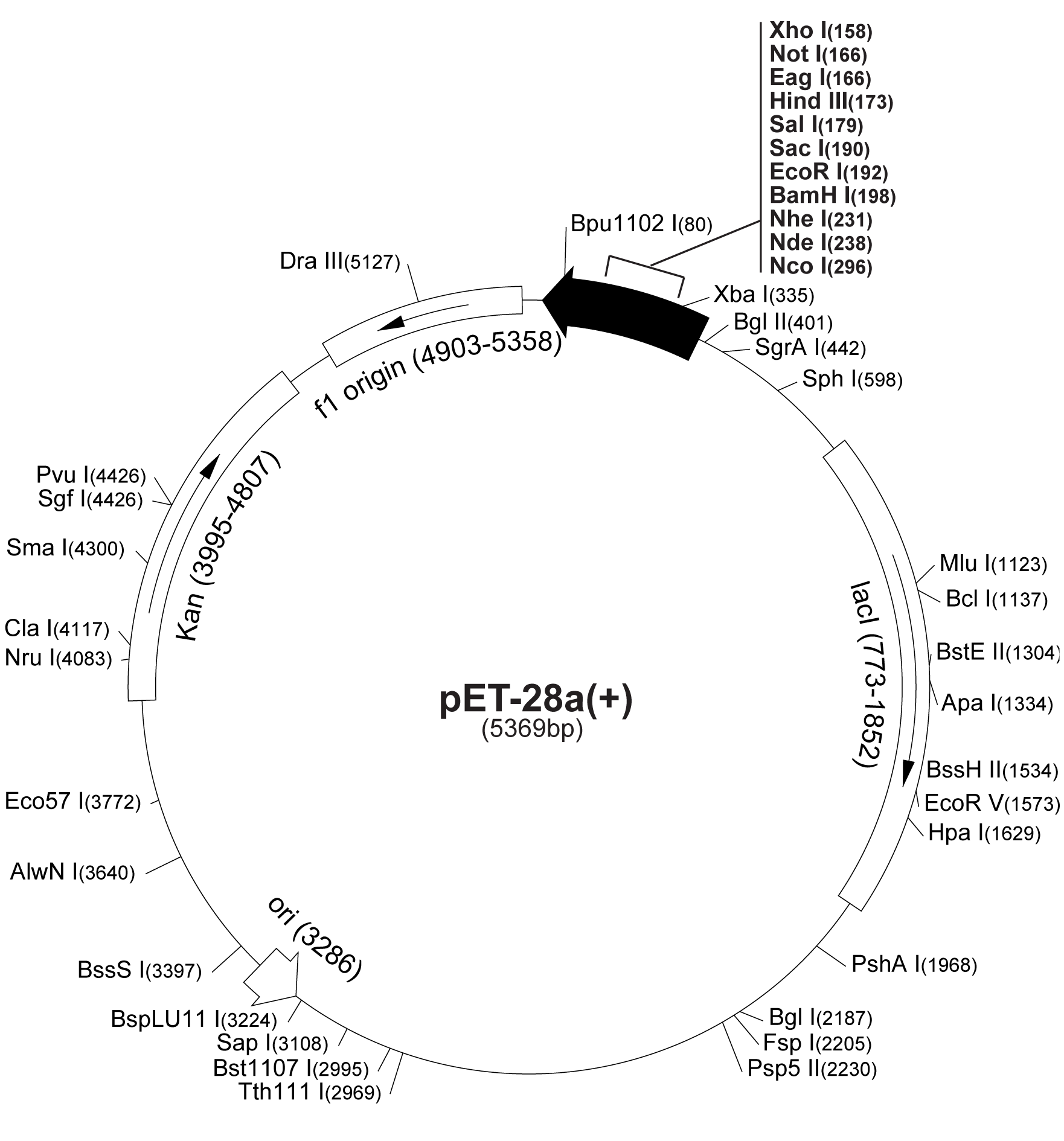
| - | [[Image: | + | [[Image:Pet28map.png|250px|thumb|Pet28 vector map]] |
|} | |} | ||
</div> | </div> | ||
Revision as of 17:01, 5 August 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
| Overall Project |
| Fuel consumption has come to the forefront as an important political and biological issue that has lead to innovative pursuits of renewable fuel as well as the controversial exploitation of natural resources. Currently ethanol is the commercial biofuel of choice; however, currently production and distillation of ethanol is inefficient. With this problem in mind, iGEM Wisconsin has looked for alternative ways to make not only ethanol, but other biofuels through synthetic biology. We've designed the following two projects using E. coli to produce biofuels in innovative ways:
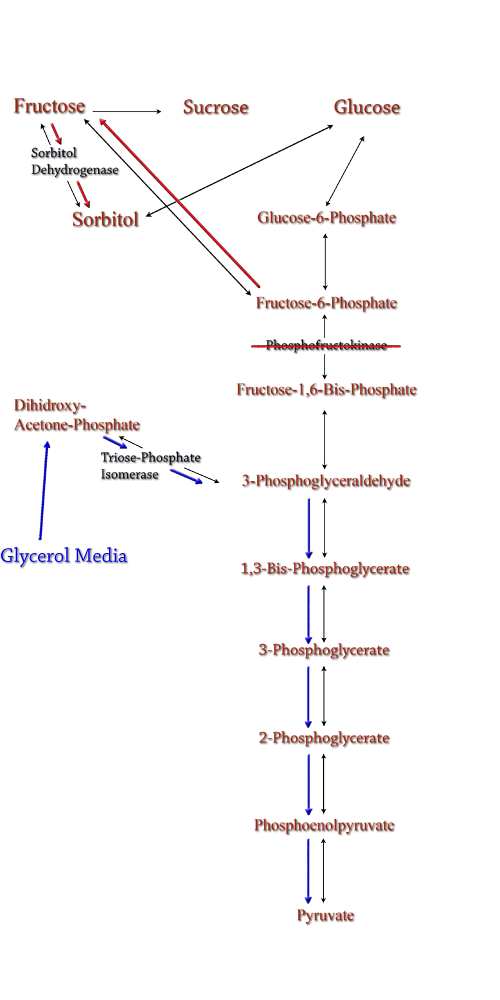
One project focuses on using E. coli to produce sorbitol, a sugar alcohol, in large quantities for eventual commercial scale catalytic conversion to hydrocarbons. Along with producing sorbitol, we've modeled alterations in E. coli to make sorbitol production from a glycerol carbon source possible. Our aim is to modify a cell that will utilize glycerol, a byproduct from biodiesel production, and effectively create sorbitol. In the second project we will be attempting to use E. coli to break down lignin from plant matter into usable biofuels. We are currently aiming to insert fungal genes coding for lignin peroxidase into E. coli. Lignin breakdown will be made possible through the transport of lignin peroxidase out of the cell. To achieve this, protein transporters will be added to the cell. |
| Sorbitol Biosynthesis |
| Sorbitol anabolism is part of the glycolysis pathway in E.coli. Interconversion between fructose and sorbitol is catalyzed by sorbitol dehydrogenase, a monomeric enzyme that uses NADH in the process. Natural expression of sorbitol dehydrogenase is generally up-regulated only in the presence of sorbitol as a carbon source. The initial idea to stimulate sorbitol over-production was to simply up-regulate the gene encoding for sorbitol dehydrogenase in an environment lacking sorbitol thus pushing the reaction governing fructose and sorbitol interconversion towards sorbitol.
|
 "
"