Team:Hawaii/Construction of Omega Interposon BioBrick
From 2008.igem.org
(Difference between revisions)
(updated methods and discussion) |
(8-14 experiment) |
||
| Line 14: | Line 14: | ||
# Transform DH5-a with the ligated product, select on Amp100. | # Transform DH5-a with the ligated product, select on Amp100. | ||
# Transform DB3.1(ccdB resistant strain) with the ligated product, select on Amp100 as a negative control. If the experimental plate has no colonies and the (-) control does, this means that the intramolecular ligation of pSB1A3 was the favored reaction. | # Transform DB3.1(ccdB resistant strain) with the ligated product, select on Amp100 as a negative control. If the experimental plate has no colonies and the (-) control does, this means that the intramolecular ligation of pSB1A3 was the favored reaction. | ||
| + | * PCR verification of the insert with VF2 and VR primers. | ||
| + | * If PCR verification is unclear, plate cells on Sm& Sp LB plates, if they grow, they have the interposon. | ||
== Results == | == Results == | ||
| - | + | :*Transformation | |
| - | * | + | ::*DH5-alpha competent cells were transformed with the ligation product (pSB1A2+omega interposon), 43 colonies were counted the next day. |
| + | ::*DB3.1 cells were also transformed with the ligation product. | ||
| + | ::*DH5-alpha cells were transformed with pUC18. | ||
| + | :*PCR verification: | ||
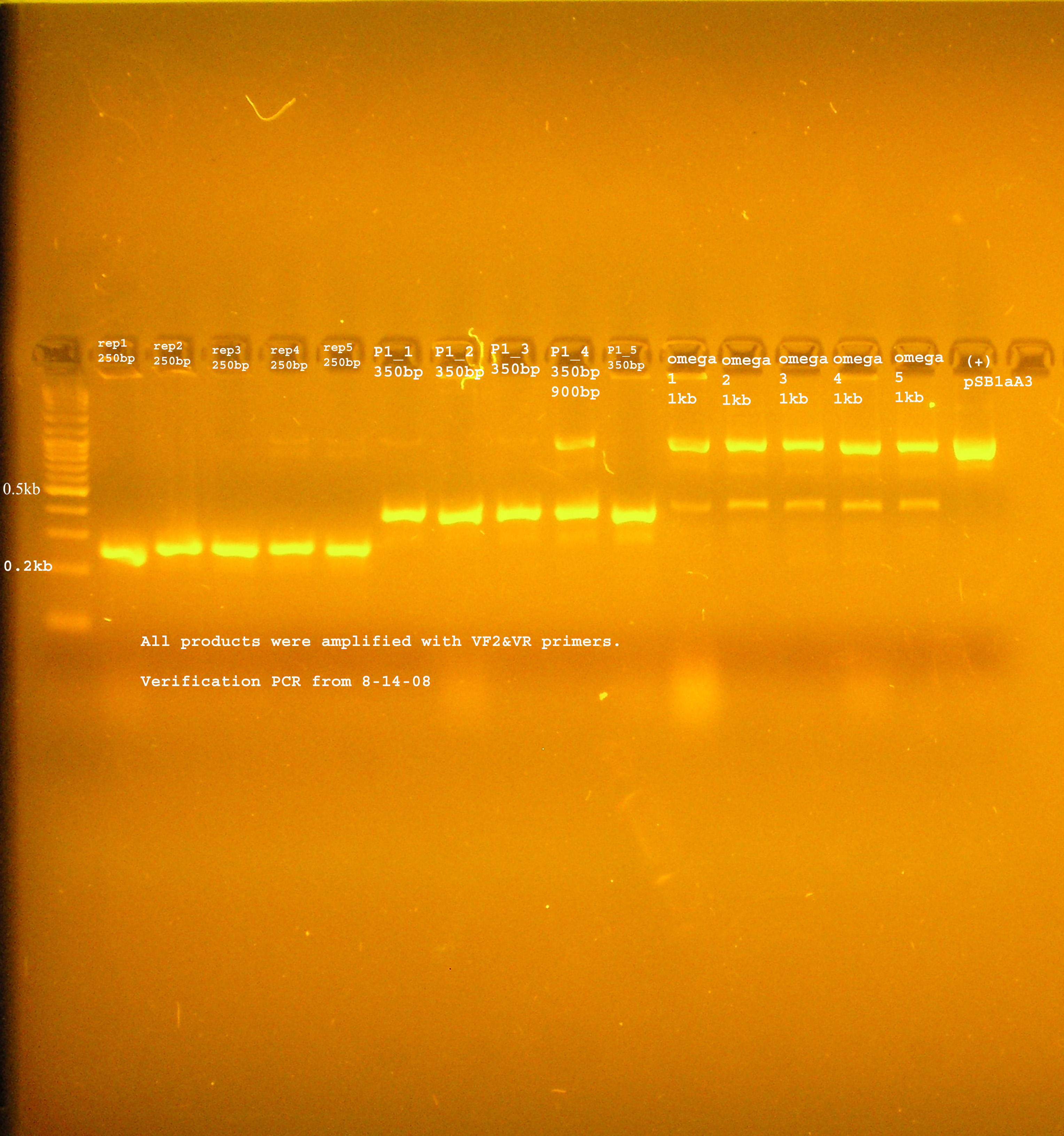
| + | ::*The bands were uniformly 1kb. | ||
| + | [[Image:omega_8_14.jpg|right|thumb|300px|PCR verification of omega interposon ligated into pSB1A2]] | ||
| + | :*Plating on Sm&Sp plates: | ||
| + | ::*No colonies grew for the ligation product | ||
| + | ::*positive control, pSMC121 was plated and a lawn of colonies grew | ||
| + | ::*negative control pSB1A2 were plated and no colonies grew | ||
== Discussion == | == Discussion == | ||
* All components of this experiment will be cut with the same restriction enzyme. This means there will probably be a large background of intramolecular ligations (pSB1A3 will ligate back to itself and pSMC121(minus the omega interposon)will ligate to itself as well). The negative control should let us know the degree to which this will occur. | * All components of this experiment will be cut with the same restriction enzyme. This means there will probably be a large background of intramolecular ligations (pSB1A3 will ligate back to itself and pSMC121(minus the omega interposon)will ligate to itself as well). The negative control should let us know the degree to which this will occur. | ||
| + | |||
| + | ===8-14 experiment=== | ||
| + | :*Transformation: | ||
| + | ::*43 colonies grew for the transformation of the ligation product into DH5-alpha cells. This means the ccdB gene was disrupted, or else the death proteins would have killed the cells. | ||
| + | ::*The transformed DB3.1 cells did not grow on ampicilin which is curious. If the ligation was successful it would have grown in both of these strains. | ||
| + | ::*That the DH5-alpha cells transformed with pUC18 did not grow makes me think there was something wrong with the transformation. | ||
| + | :*PCR verification | ||
| + | ::*The band was consistently around 1kb. This is the size expected if the ccdB region was amplified. | ||
| + | :*Sm&Sp plating | ||
| + | ::*It is clear that the cells transformed with the ligation product do not contain a functional version of the omega interposon because the cells showed no resistance to the antibiotics. | ||
| + | |||
| + | :*Conclusions: | ||
| + | ::*I need to run a gel to verify that the omega interposon was digested properly away from its vector. This digestion product can be purified from the gel and another ligation can be performed. | ||
{{Team:Hawaii/Footer}} | {{Team:Hawaii/Footer}} | ||
Revision as of 00:04, 17 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
Construction of Omega Interposon BioBrick
- The omega interposon (OI) BioBrick will be constructed using insertional mutagenesis methods. OI will be inserted into the ccdB region of pSB1A3 by restricting both pSB1A3 (containing ccdB) and pSMC121-a plasmid containing OI- with either XmaI or SmaI, depending on which enzyme is more available.
- The omega interposon is an insertional mutagenesis tool containing the aadA gene from plasmid R1001.1 which confers Spectinomycin and Streptomycin resistance. Flanking the aadA gene are transcriptional termination sites of the T4 gene 32 so that transcription cannot be achieved through the omega interposon from either side. To avoid polypeptide synthesis at the position of the omega interposon, synthetic translational stop codons were also included. Flanking this feature are two polylinkers (Prentki 1983). OI can be used in experiments where disruption of a gene is needed. These experiments are often performed when discerning the function of a gene.
Methods
- Cut pSMC121 with SmaI or XmaI; cut pSB1A3 with the same enzyme.
- Purify restriction digests to get rid of small digest fragments.
- Ligate the omega interposon with pSB1A3
- Transformation:
- Transform DH5-a with the ligated product, select on Amp100.
- Transform DB3.1(ccdB resistant strain) with the ligated product, select on Amp100 as a negative control. If the experimental plate has no colonies and the (-) control does, this means that the intramolecular ligation of pSB1A3 was the favored reaction.
- PCR verification of the insert with VF2 and VR primers.
- If PCR verification is unclear, plate cells on Sm& Sp LB plates, if they grow, they have the interposon.
Results
- Transformation
- DH5-alpha competent cells were transformed with the ligation product (pSB1A2+omega interposon), 43 colonies were counted the next day.
- DB3.1 cells were also transformed with the ligation product.
- DH5-alpha cells were transformed with pUC18.
- PCR verification:
- The bands were uniformly 1kb.
- Plating on Sm&Sp plates:
- No colonies grew for the ligation product
- positive control, pSMC121 was plated and a lawn of colonies grew
- negative control pSB1A2 were plated and no colonies grew
Discussion
- All components of this experiment will be cut with the same restriction enzyme. This means there will probably be a large background of intramolecular ligations (pSB1A3 will ligate back to itself and pSMC121(minus the omega interposon)will ligate to itself as well). The negative control should let us know the degree to which this will occur.
8-14 experiment
- Transformation:
- 43 colonies grew for the transformation of the ligation product into DH5-alpha cells. This means the ccdB gene was disrupted, or else the death proteins would have killed the cells.
- The transformed DB3.1 cells did not grow on ampicilin which is curious. If the ligation was successful it would have grown in both of these strains.
- That the DH5-alpha cells transformed with pUC18 did not grow makes me think there was something wrong with the transformation.
- PCR verification
- The band was consistently around 1kb. This is the size expected if the ccdB region was amplified.
- Sm&Sp plating
- It is clear that the cells transformed with the ligation product do not contain a functional version of the omega interposon because the cells showed no resistance to the antibiotics.
- Conclusions:
- I need to run a gel to verify that the omega interposon was digested properly away from its vector. This digestion product can be purified from the gel and another ligation can be performed.
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"