Team:Hawaii/Notebook/2008-08-11
From 2008.igem.org
(Difference between revisions)
(add margarets experiments) |
(→Construction of GFP device) |
||
| Line 63: | Line 63: | ||
!DNA concentration | !DNA concentration | ||
|- | |- | ||
| - | |nir+rbs | + | |align=center|nir+rbs |
| - | |4.8 ng/μl | + | |align=center|4.8 ng/μl |
|- | |- | ||
| - | |plac+rbs | + | |align=center|plac+rbs |
| - | |3.6 ng/μl | + | |align=center|3.6 ng/μl |
|- | |- | ||
| - | |GFP | + | |align=center|GFP |
| - | |4.7 ng/μl | + | |align=center|4.7 ng/μl |
|- | |- | ||
| - | |GFPfusion | + | |align=center|GFPfusion |
| - | |6.4 ng/μl | + | |align=center|6.4 ng/μl |
|} | |} | ||
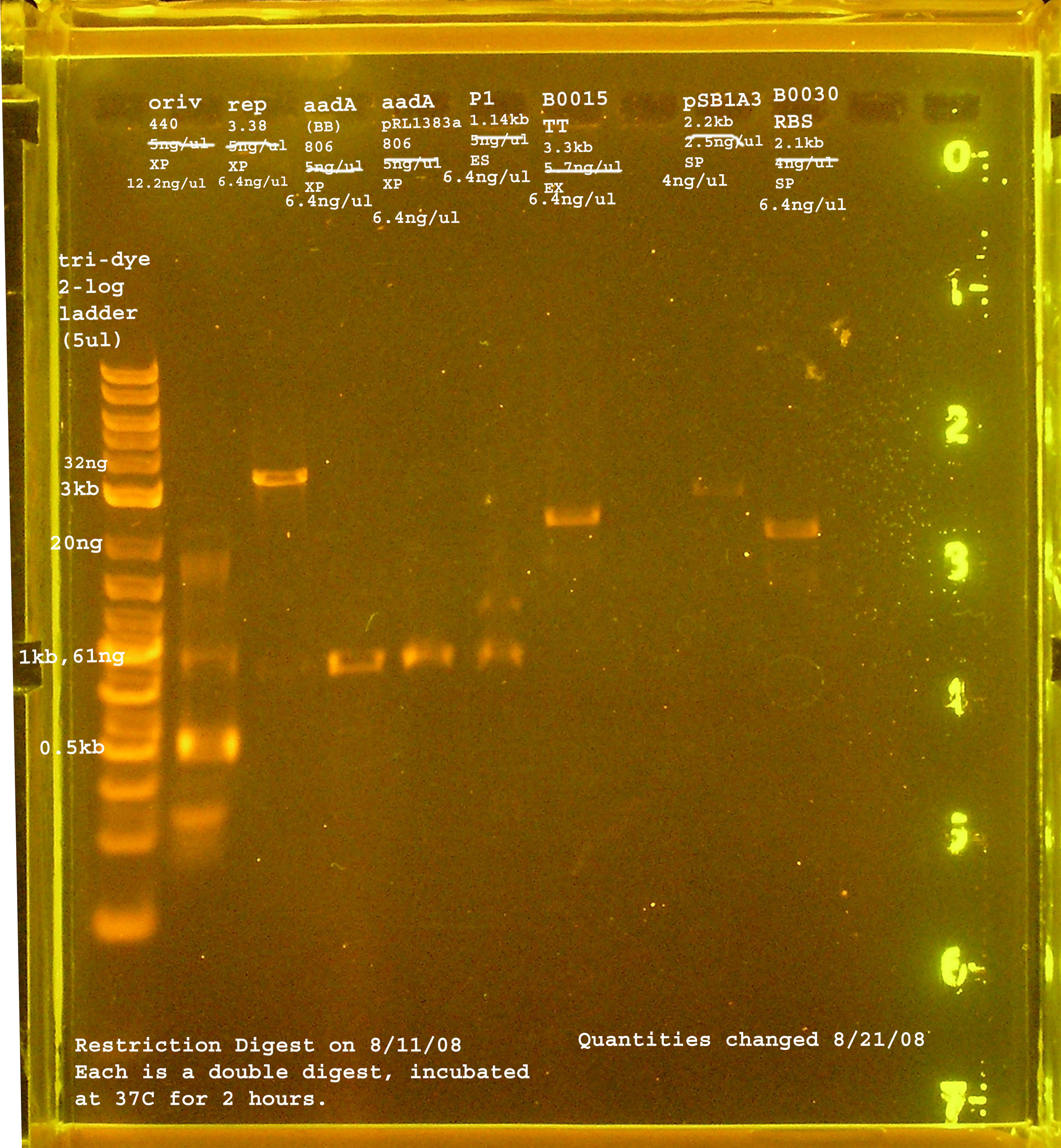
| + | [[Image:081108REdigests.jpg|right|thumb|200px|EtBr stained 2% agarose gel ran at 72V for 1.5 hours. Thirty microliters of the RE digest reactions were loaded into each well.]] | ||
:* Restriction digested in 30 μl reactions: | :* Restriction digested in 30 μl reactions: | ||
::* B0015 with XbaI then EcoRI | ::* B0015 with XbaI then EcoRI | ||
::* GFP and GFPf with EcoRI and SpeI | ::* GFP and GFPf with EcoRI and SpeI | ||
::* slr1, slr2, pilA with SpeI and PstI | ::* slr1, slr2, pilA with SpeI and PstI | ||
| - | :* Ran new RE digests EtBr stained 2% agarose gel at 72V for | + | :* Ran new RE digests EtBr stained 2% agarose gel at 72V for 1.5 hours |
:* Extracted parts from gel and determined DNA concentrations | :* Extracted parts from gel and determined DNA concentrations | ||
| + | {|class=wikitable border=1 align=center | ||
| + | !Part | ||
| + | !DNA concentration | ||
| + | |- | ||
| + | |align=center|slr1 | ||
| + | |align=center|2.6 ng/μl | ||
| + | |- | ||
| + | |align=center|pilA | ||
| + | |align=center| 1.1 ng/μl | ||
| + | |- | ||
| + | |align=center|GFP | ||
| + | |align=center|0.4 ng/μl | ||
| + | |- | ||
| + | |align=center|GFPf | ||
| + | |align=center|11.3 ng/μl | ||
| + | |- | ||
| + | |align=center|B0015 | ||
| + | |align=center|1.9 ng/μl | ||
| + | |} | ||
| + | :* Ligated: | ||
| + | ::* 8 μl GFP + 0.5 μl B0015 | ||
| + | ::* 4 μl GFPf + 4 7mu;l B0015 | ||
| + | ::* 2 μl GFPf + 1.5 μl slr1 | ||
| + | ::* 2 μl GFPf + 3.5 μl pilA | ||
| + | :* Transformed 7 μl ligation reaction into DB3.1 cells | ||
| + | :* RE digest overnight of 22 μl pSB1A2 with EcoRI and PstI for 3A assembly | ||
| + | |||
| + | ===Testing restriction enzymes in the lab's -20C freezer=== | ||
| + | :<strong>Grace</strong> | ||
| + | |||
| + | :* Digested pRL1383a with BamHI (should result in a single linear fragment) | ||
| + | :* Digested pRL1383a with HindIII (should result in a single linear fragment) | ||
| + | :* Digested plasmid preps (E0240, I14032, I51020, nir+rbs, plac+rbs) with NotI (should result in two fragments -- vector and insert) | ||
===[[Team:Hawaii/Ligation of pRL1383a Parts|Ligation of pRL1383a Parts]]=== | ===[[Team:Hawaii/Ligation of pRL1383a Parts|Ligation of pRL1383a Parts]]=== | ||
Revision as of 03:51, 12 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
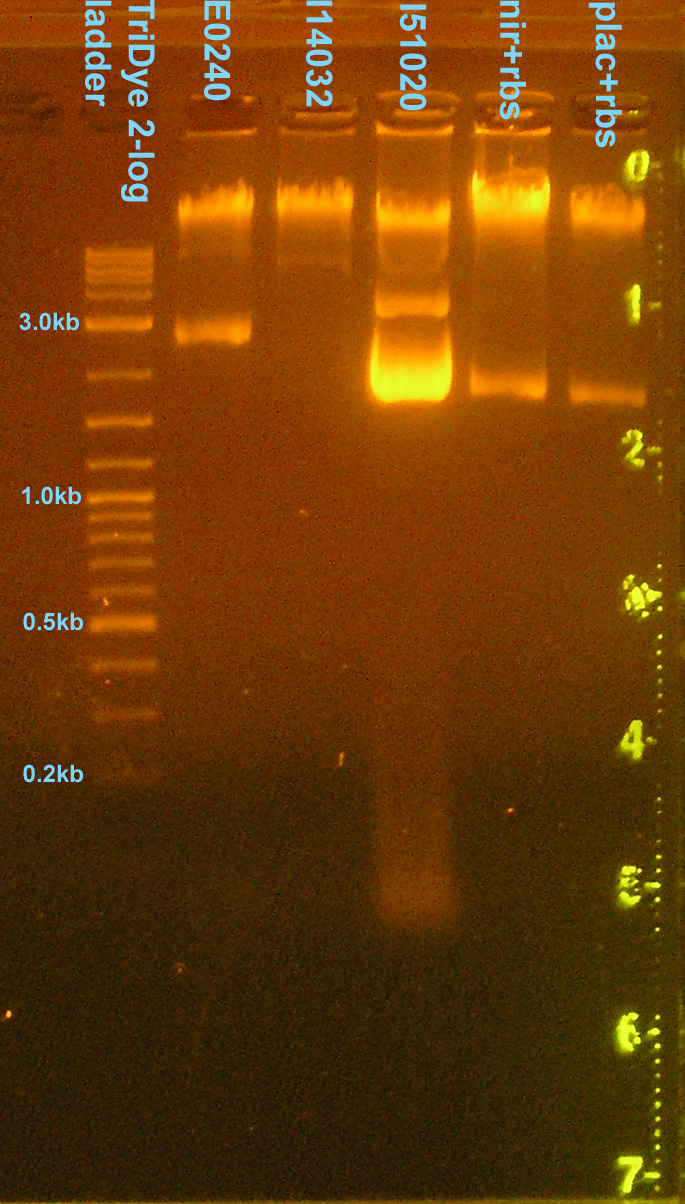
Checked plasmid prep from weekend
- Grace
- Ran on 2.0% agarose gel to verify plasmids
- DNA didn't run. Agarose concentration too high. Redid on 0.8% gel.
- Genomic DNA up top?
- Clean prep (no RNA)!
- Only E0240 verified. All other bands wrong size (circular/supercoiled?)
- Checked DNA concentrations via nanodrop spectrometer
| Plasmid | DNA concentration | 260/280 | 260/230 |
|---|---|---|---|
| E0240 | 757.7 ng/μl | 2.06 | 1.49 |
| I14032 (2005 distribution) | 541.4 ng/μl | 2.01 | 1.27 |
| I51020 | 2775.6 ng/μl | 1.97 | 1.77 |
| nir+rbs | 566.8 ng/μl | 1.83 | 1.10 |
| plac+rbs | 344.0 ng/μl | 1.95 | 1.28 |
Made 1000x Amp,100 stock solution
- Grace
Reinoculated for cryostocking
- Grace
- I14032 from 2005 and 2008 distributions
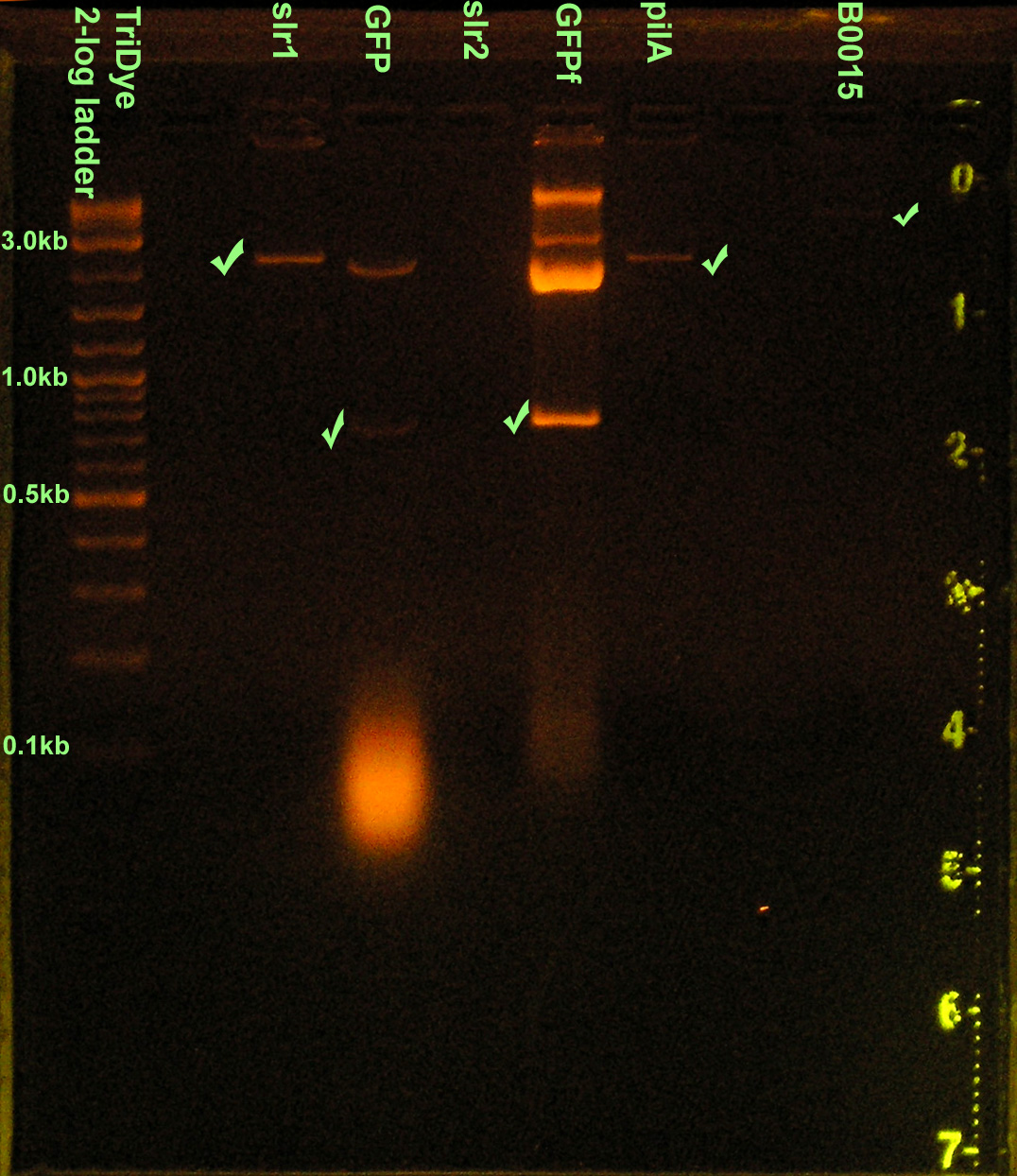
Construction of GFP device
- Grace
- Extracted nir+rbs, plac+rbs, GFP, GFPf from gel ran yesterday
- B0015 could not be extracted because fragment was not visible under short wave UV
- Digestion was done for 3A assembly rather than rear ligation (oops). Redid RE digest.
- Checked DNA concentrations via nanodrop spectrometer
| Part | DNA concentration |
|---|---|
| nir+rbs | 4.8 ng/μl |
| plac+rbs | 3.6 ng/μl |
| GFP | 4.7 ng/μl |
| GFPfusion | 6.4 ng/μl |
- Restriction digested in 30 μl reactions:
- B0015 with XbaI then EcoRI
- GFP and GFPf with EcoRI and SpeI
- slr1, slr2, pilA with SpeI and PstI
- Ran new RE digests EtBr stained 2% agarose gel at 72V for 1.5 hours
- Extracted parts from gel and determined DNA concentrations
| Part | DNA concentration |
|---|---|
| slr1 | 2.6 ng/μl |
| pilA | 1.1 ng/μl |
| GFP | 0.4 ng/μl |
| GFPf | 11.3 ng/μl |
| B0015 | 1.9 ng/μl |
- Ligated:
- 8 μl GFP + 0.5 μl B0015
- 4 μl GFPf + 4 7mu;l B0015
- 2 μl GFPf + 1.5 μl slr1
- 2 μl GFPf + 3.5 μl pilA
- Transformed 7 μl ligation reaction into DB3.1 cells
- RE digest overnight of 22 μl pSB1A2 with EcoRI and PstI for 3A assembly
Testing restriction enzymes in the lab's -20C freezer
- Grace
- Digested pRL1383a with BamHI (should result in a single linear fragment)
- Digested pRL1383a with HindIII (should result in a single linear fragment)
- Digested plasmid preps (E0240, I14032, I51020, nir+rbs, plac+rbs) with NotI (should result in two fragments -- vector and insert)
Ligation of pRL1383a Parts
Margaret
- restriction digest of rep, oriV, aada(BB), aada(pRL1383a), P1 lytic region, pSB1A3, B0030, B0015
- Ligation: rep+B0030, oriV+pSB1A3, aadA(BB)+B0030, aadA(pRL1383a)+B0030, P1 lytic + B0015, pSB1A3 to itself (-) control
- Transformation into DH5-a (batch 3)
Started Culture for plasmid prep & cryostocks
- to be completed 8/12
- B0015, pSB3K3, oriT(cryostock & plasmid prep), B0030, I14032, E0040, J33207
Discussion
- FYI:
- According to the Endy lab, ligation reactions should have <100ng DNA per reaction for maximum efficiency
- ~10ng vector should be used in ligation reactions (6:1 ratio of insert to vector)
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"