Team:Paris/Analysis/Design2
From 2008.igem.org
(→Limits of our network) |
(→Implementation of the core system) |
||
| Line 25: | Line 25: | ||
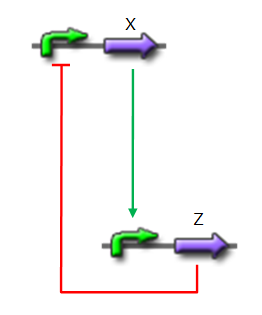
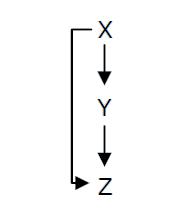
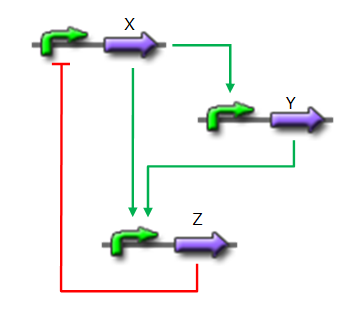
*X is flhDC, the master regulator of the synthesis of the flagella. It is associated to its natural promoter. | *X is flhDC, the master regulator of the synthesis of the flagella. It is associated to its natural promoter. | ||
*Y is fliA, another transcription factor that regulates the expression of a large amount of flagellar genes. fliA is also associated with its natural promoter. | *Y is fliA, another transcription factor that regulates the expression of a large amount of flagellar genes. fliA is also associated with its natural promoter. | ||
| - | *For Z, we need a protein that inhibits the expression of flhDC. [http://www.ncbi.nlm.nih.gov/pubmed/18193944?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Antoine Giraud ''et al.''(2008)] | + | *For Z, we need a protein that inhibits the expression of flhDC. |
| + | We considered two possibilities. First of all, we planned to use tetR, and the appropriate promoter before flhDC. But, once again, in a concern to use as many "natural" BioBricks, we decided to use proteins that naturally inhibits the expression of flhDC, such as envZ and ompR. | ||
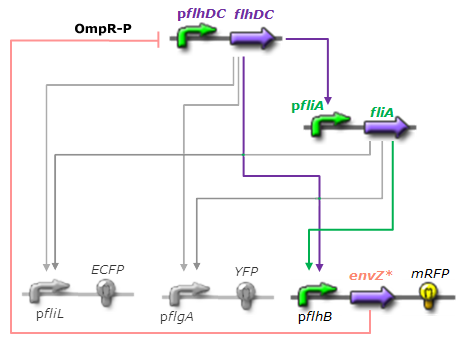
| + | The mechanism of inhibition is quite well-known. envZ phosphorylates OmpR which becomes active. OmpR-P strongly inhibits the expression of flhDC. [http://www.ncbi.nlm.nih.gov/pubmed/18193944?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Antoine Giraud ''et al.''(2008)] discribed a specific mutant, envZ* that phosphorylates very efficiently ompR. We chose envZ* for Z because it introduces another delay, as envZ* works through over-phophorylation of ompR, which in turns represses flhDC. For Z promoter, we chose one of the promoter controlling the expression of one of the flagellar gene that are regulated by both FliA and FlhDC. We chose the promoter of FlhB because it is the gene that is lastly activated. As a consequence, it increases the length of each cycle. Of course, to make the oscillations observable, we decided to put EnvZ and GFP under the control of the same promoter. | ||
[[Image:Reseauoscillfinal.png|center|thumb|250 px|Network 3. Final Design of the core system.]] | [[Image:Reseauoscillfinal.png|center|thumb|250 px|Network 3. Final Design of the core system.]] | ||
Revision as of 17:57, 28 October 2008
|
Network Design - Part 2
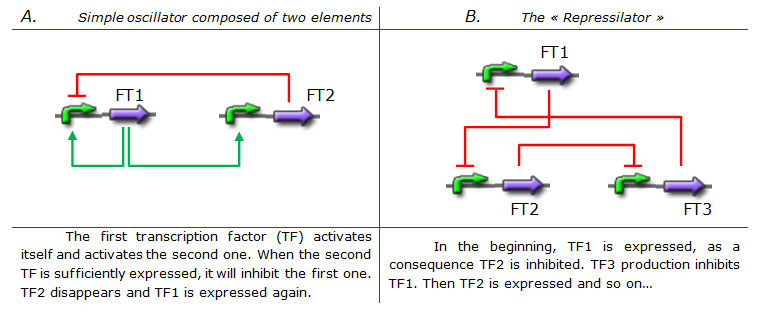
Creating an oscillating systemAlready existing genetic oscillators and their limitsDesigning a simple genetic network that presents an oscillatory behavior is one of the first challenge synthetic biology overcame. More or less successfully. We can count more than ten synthetic genetic oscillators that have varied period and mechanisms. [http://www.ncbi.nlm.nih.gov/pubmed/16604190?ordinalpos=2&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Raúl GUANTES and Juan F. POYATOS (2006)] studied the most simple oscillators composed of two elements while [http://www.ncbi.nlm.nih.gov/pubmed/10659856?ordinalpos=10&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Michael B. ELOWITZ and Stanislas LEIBLER (2000)] designed the more complex "repressilator" (Table 1), to quote only the best known. Both oscillators work : we can observe oscillations but only a limited number of cycles. Actually, they always reach a steady-state because the degradation/dilution rate is often too low : at the end of each cycle, the conditions are not exactly the initial conditions. Experimentally, the longer is the period the more cycles we can observe. Design of our genetic oscillator : The Feed Forward LoopWe want to design a simple oscillator that oscillates during as many cycles as possible. We propose a system based on an oscillator composed of two elements (Network 1) on which we added a delay at the end of each cycle. Uri ALON described genetic network motifs that generate a delay. Those motifs are the type 1 coherent Feed Forward Loop (C1-FFL). ↓ More on Feed Forward Loop ↑
We will use one of those network to increase the run of each period and permit more oscillations (Network 2). Implementation of the core system[http://www.ncbi.nlm.nih.gov/pubmed/16729041?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Shiraz Kalir et al. (2004)] studied the complex network of gene that lead to the synthesis of E. coli flagella. A C1-FFL is present in this network.
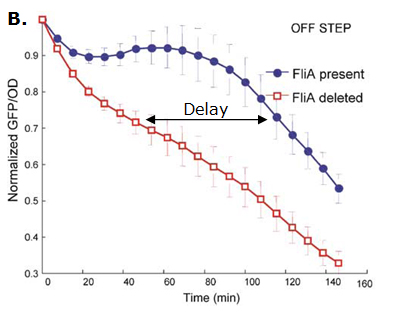
We considered two possibilities. First of all, we planned to use tetR, and the appropriate promoter before flhDC. But, once again, in a concern to use as many "natural" BioBricks, we decided to use proteins that naturally inhibits the expression of flhDC, such as envZ and ompR. The mechanism of inhibition is quite well-known. envZ phosphorylates OmpR which becomes active. OmpR-P strongly inhibits the expression of flhDC. [http://www.ncbi.nlm.nih.gov/pubmed/18193944?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum Antoine Giraud et al.(2008)] discribed a specific mutant, envZ* that phosphorylates very efficiently ompR. We chose envZ* for Z because it introduces another delay, as envZ* works through over-phophorylation of ompR, which in turns represses flhDC. For Z promoter, we chose one of the promoter controlling the expression of one of the flagellar gene that are regulated by both FliA and FlhDC. We chose the promoter of FlhB because it is the gene that is lastly activated. As a consequence, it increases the length of each cycle. Of course, to make the oscillations observable, we decided to put EnvZ and GFP under the control of the same promoter. Limits of our networkIntuitively, it seems that there is a range of parameters that permit oscillations. However, an analysis of the core system highlighted the fact that it could hardly have an oscillating dynamics (Graph 1). Among the alternatives we studied, the system that could most probably oscillate is the third module that uses the quorum sensing to produce both a delay and the synchronization at the population level.
|
 "
"