Team:Cambridge/Voltage
From 2008.igem.org
| Line 20: | Line 20: | ||
'''In order to simulate neural activity in bacteria, a mechanism resembling a synapse is necessary. This is the aim of the Voltage section of the Cambridge iBRAIN project. At the synapse, neurotransmitter molecules are released from the presynaptic plasma membrane. The neurotransmitter diffuses through the synaptic cleft and binds to chemical receptor molecules on the membrane of the postsynaptic cell. These receptors cause ion channels to open so that ions rush in or out, changing the transmembrane potential. Attempting to mimic this in a prokaryotic system is particularly attractive as, in a more general sense, it provides an interface between chemical or biological and electrical systems.''' | '''In order to simulate neural activity in bacteria, a mechanism resembling a synapse is necessary. This is the aim of the Voltage section of the Cambridge iBRAIN project. At the synapse, neurotransmitter molecules are released from the presynaptic plasma membrane. The neurotransmitter diffuses through the synaptic cleft and binds to chemical receptor molecules on the membrane of the postsynaptic cell. These receptors cause ion channels to open so that ions rush in or out, changing the transmembrane potential. Attempting to mimic this in a prokaryotic system is particularly attractive as, in a more general sense, it provides an interface between chemical or biological and electrical systems.''' | ||
| - | [[Image:GluR0.PNG|right| | + | [[Image:GluR0.PNG|right|370px|GluR0 Plasmid]] |
Using the iGEM synthetic biology concept, we identified genes which could contribute to a synapse-like response. We also used a number of existing | Using the iGEM synthetic biology concept, we identified genes which could contribute to a synapse-like response. We also used a number of existing | ||
[[IGEM:Cambridge/2008/Extracted_Parts | parts from the registry]]. To keep the system as simple and therefore feasible as possible the aim was to engineer E.coli to respond to a specific ligand by allowing a flux of ions which would be measurable as a small change in the voltage of the medium the cells are suspended in. The first step was to find prokaryotic ion channels which open or close in response to a particular molecule or small set of molecules. We searched the [http://www.uniprot.org/ Uniprot database] and found that the organism [http://www.ebi.ac.uk/2can/genomes/bacteria/Synechocystis.html Synechocystis PCC 6803] has a transmembrane protein, GluR0 which is glutamate-gated, and allows flux of potassium ions when opened. We decided to try using the protein coding sequence of the gene for this channel. However, as the sequence came from a different organism, it was necessary to optimise the codons for expression in E.coli. We therefore decided to have the [[IGEM:Cambridge/2008/Notebook/Voltage/GluR0 Manipulation| GluRo Biobrick plasmid]] we had designed synthesised by DNA2.0. [[IGEM:Cambridge/2008/Notebook/Voltage/Gene Design|''Gene Design'']] | [[IGEM:Cambridge/2008/Extracted_Parts | parts from the registry]]. To keep the system as simple and therefore feasible as possible the aim was to engineer E.coli to respond to a specific ligand by allowing a flux of ions which would be measurable as a small change in the voltage of the medium the cells are suspended in. The first step was to find prokaryotic ion channels which open or close in response to a particular molecule or small set of molecules. We searched the [http://www.uniprot.org/ Uniprot database] and found that the organism [http://www.ebi.ac.uk/2can/genomes/bacteria/Synechocystis.html Synechocystis PCC 6803] has a transmembrane protein, GluR0 which is glutamate-gated, and allows flux of potassium ions when opened. We decided to try using the protein coding sequence of the gene for this channel. However, as the sequence came from a different organism, it was necessary to optimise the codons for expression in E.coli. We therefore decided to have the [[IGEM:Cambridge/2008/Notebook/Voltage/GluR0 Manipulation| GluRo Biobrick plasmid]] we had designed synthesised by DNA2.0. [[IGEM:Cambridge/2008/Notebook/Voltage/Gene Design|''Gene Design'']] | ||
| Line 28: | Line 28: | ||
The idea of being able to measure a change in voltage of the medium brings with it a number of challenges. The number of ions flowing across the membrane should be as high as possible, to produce a signal significantly higher than the noise. This could be achieved by first packing the cells with potassium, so that a large gradient was set up between the inside and the outside of the cell. Then when the GluR0 channels open, potassium ions will suddenly flow down the concentration gradient. In order to build up this gradient in the first place, we planned to over-express the Kdp-ATPase, which would pump potassium into the cell. However, to stop the balance being restored, the potassium channels would need to be absent or disabled. | The idea of being able to measure a change in voltage of the medium brings with it a number of challenges. The number of ions flowing across the membrane should be as high as possible, to produce a signal significantly higher than the noise. This could be achieved by first packing the cells with potassium, so that a large gradient was set up between the inside and the outside of the cell. Then when the GluR0 channels open, potassium ions will suddenly flow down the concentration gradient. In order to build up this gradient in the first place, we planned to over-express the Kdp-ATPase, which would pump potassium into the cell. However, to stop the balance being restored, the potassium channels would need to be absent or disabled. | ||
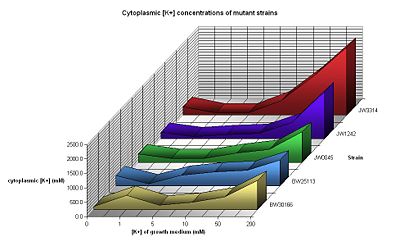
| - | [[Image:Kallstrains.JPG|left| | + | [[Image:Kallstrains.JPG|left|350px|K+ Concentrations]] |
We ordered a variety of [[IGEM:Cambridge/2008/Notebook/Voltage/Mutant Strains |mutant E.coli strains]] with mutations in each of the genes coding for the potassium flux membrane proteins Kdp, Kch and Kef. To examine the behaviour of these mutant strains we grew them in a variety of potassium concentrations and used [[IGEM:Cambridge/2008/Notebook/Voltage/OD600 Calibration| OD600 readings]] to measure their [[IGEM:Cambridge/2008/Notebook/Voltage/K+ Growth|growth rates]]. We also measured [[IGEM:Cambridge/2008/Notebook/Voltage/K+ Concentrations| internal potassium concentrations]] using [[IGEM:Cambridge/2008/Notebook/Voltage/Flame Photometer Calibration|flame photometry]]. Unfortunately there was little difference seen between most of the mutants and the control strain, and flame photometry readings were not always reliable. | We ordered a variety of [[IGEM:Cambridge/2008/Notebook/Voltage/Mutant Strains |mutant E.coli strains]] with mutations in each of the genes coding for the potassium flux membrane proteins Kdp, Kch and Kef. To examine the behaviour of these mutant strains we grew them in a variety of potassium concentrations and used [[IGEM:Cambridge/2008/Notebook/Voltage/OD600 Calibration| OD600 readings]] to measure their [[IGEM:Cambridge/2008/Notebook/Voltage/K+ Growth|growth rates]]. We also measured [[IGEM:Cambridge/2008/Notebook/Voltage/K+ Concentrations| internal potassium concentrations]] using [[IGEM:Cambridge/2008/Notebook/Voltage/Flame Photometer Calibration|flame photometry]]. Unfortunately there was little difference seen between most of the mutants and the control strain, and flame photometry readings were not always reliable. | ||
| Line 48: | Line 48: | ||
[http://www.springerlink.com/content/6042632827845551/ The Kdp-ATPase system and its regulation] | [http://www.springerlink.com/content/6042632827845551/ The Kdp-ATPase system and its regulation] | ||
| - | + | [http://cgsc.biology.yale.edu/Strain.php?ID=107402 Potential Chassis: Strain JW1242-1] | |
[http://cgsc.biology.yale.edu/Strain.php?ID=107065 Strain JW0710-1] | [http://cgsc.biology.yale.edu/Strain.php?ID=107065 Strain JW0710-1] | ||
Revision as of 22:05, 28 October 2008
|
||
 "
"