Team:LCG-UNAM-Mexico/Notebook/2008-August
From 2008.igem.org
(Difference between revisions)
| Line 125: | Line 125: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"><div align="justify"><p><strong | + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong><br></p> |
<p>We left PCR of:</p> | <p>We left PCR of:</p> | ||
<ul> | <ul> | ||
| Line 141: | Line 141: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"><div align="justify"><p><b | + | <td class="bodyText"><div align="justify"><p><b>MODELING:</b><br>Hill cooperativity 5th Reaction Reminder: </p> |
<p>A + B <--> AB <br /> | <p>A + B <--> AB <br /> | ||
<strong>Ka=Keq=[AB]/[A][B]=1/Kd</strong> <br /> | <strong>Ka=Keq=[AB]/[A][B]=1/Kd</strong> <br /> | ||
θ=[AB]/([AB]+[A])=[B]/([B]+Kd) </p><br> | θ=[AB]/([AB]+[A])=[B]/([B]+Kd) </p><br> | ||
| - | <p><strong>MWC Model</strong> (Cooperativity) <br /> | + | <p><strong><u>MWC Model</u></strong> (Cooperativity) <br /> |
A + nB <--> ABn <br /> | A + nB <--> ABn <br /> | ||
<strong>Ka=Keq=[ABn]/[A][B]n=1/Kd</strong> <br /> | <strong>Ka=Keq=[ABn]/[A][B]n=1/Kd</strong> <br /> | ||
| Line 198: | Line 198: | ||
</table> | </table> | ||
<p> </p> | <p> </p> | ||
| - | <p><strong | + | <p><strong>WET LAB:</strong></p> |
| - | <p><strong>Gel</strong></p> | + | <p><strong><u>Gel</u></strong></p> |
<p>We run a gel with the PCR products obtained the day 01-08-08</p> | <p>We run a gel with the PCR products obtained the day 01-08-08</p> | ||
| - | <p><strong>Purification</strong></p> | + | <p><strong><u>Purification</u></strong></p> |
<p>From the PCR of the 01-08-08 we took 120 μl to purify DNA in a low fusion point agarose gel in line with the kit.</p> | <p>From the PCR of the 01-08-08 we took 120 μl to purify DNA in a low fusion point agarose gel in line with the kit.</p> | ||
<p>We run an agarose gel to verify the status of the purified PRC products.</p> | <p>We run an agarose gel to verify the status of the purified PRC products.</p> | ||
| Line 213: | Line 213: | ||
<li> Operator of rcnR + RcnA</li> | <li> Operator of rcnR + RcnA</li> | ||
</ol> | </ol> | ||
| - | <p><strong>Restrictions</strong><br /> | + | <p><u><strong>Restrictions</strong></u><br /> |
</p> | </p> | ||
<p>We left restrictions over night(double and simple) of each biopart.</p> | <p>We left restrictions over night(double and simple) of each biopart.</p> | ||
| Line 280: | Line 280: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"> <div align="justify"><p><b | + | <td class="bodyText"> <div align="justify"><p><b>MODELING:</b><br>Hill Cooperativity<br /> |
</p> | </p> | ||
<p>5th Reaction, conflict<br /> | <p>5th Reaction, conflict<br /> | ||
| Line 294: | Line 294: | ||
<p> The proposed explanation is that the equation used to determine k + does not fit our model. We should explore other possibilities. </p> | <p> The proposed explanation is that the equation used to determine k + does not fit our model. We should explore other possibilities. </p> | ||
<p> </p> | <p> </p> | ||
| - | <p><strong | + | <p><strong>WET LAB:</strong></p> |
| - | <p><strong>Restrictions</strong></p> | + | <p><strong><u>Restrictions</u></strong></p> |
<p><strong>Second simple restriction</strong></p> | <p><strong>Second simple restriction</strong></p> | ||
<p>Before the second simple restriction we cleaned the product oof the first restriction with the purification Kit.<br /> | <p>Before the second simple restriction we cleaned the product oof the first restriction with the purification Kit.<br /> | ||
| Line 348: | Line 348: | ||
</li> | </li> | ||
</ul> | </ul> | ||
| - | <p><strong>Extraction</strong></p> | + | <p><strong><u>Extraction</u></strong></p> |
<p>Plasmids pRK415 and pBBR1MCS-5 were extracted with the Roche kit(see Techniques).</p> | <p>Plasmids pRK415 and pBBR1MCS-5 were extracted with the Roche kit(see Techniques).</p> | ||
| - | <p><strong>Cultures</strong></p> | + | <p><strong><u>Cultures</u></strong></p> |
<p>We cultured DH5alfa cells tranfromed with pJet+biopart</p> | <p>We cultured DH5alfa cells tranfromed with pJet+biopart</p> | ||
| Line 359: | Line 359: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"><div align="justify"><p><b | + | <td class="bodyText"><div align="justify"><p><b>MODELING:</b><br>Hill Cooperativity: <br /> |
5th Reaction, solving the problem: <br /> | 5th Reaction, solving the problem: <br /> | ||
<br /> | <br /> | ||
| Line 378: | Line 378: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"><div align="justify"><p><strong | + | <td class="bodyText"><div align="justify"><p><strong>GROUP MEETING </strong><br /> |
| - | Wet Lab | + | Wet Lab Statusk<br /> |
<strong><br /> | <strong><br /> | ||
Objectives: </strong><br /> | Objectives: </strong><br /> | ||
| Line 416: | Line 416: | ||
</div></td> | </div></td> | ||
</tr> | </tr> | ||
| + | |||
<tr> | <tr> | ||
<td class="subHeader" bgcolor="#99CC66" id="12">2008-08-12</td> | <td class="subHeader" bgcolor="#99CC66" id="12">2008-08-12</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td class="bodyText"><div align="justify"><p><strong | + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong></p> |
| - | <p><strong>Cultures</strong></p> | + | <p><u><strong>Cultures</strong></u></p> |
<p>We left cultures of Biopart 1 in pJEt</p> | <p>We left cultures of Biopart 1 in pJEt</p> | ||
| - | <p><strong>Plasmid Extraction</strong></p> | + | <p><strong><u>Plasmid Extraction</u></strong></p> |
<p>Plasmid RcnA was extracted by alkaline lysis</p> | <p>Plasmid RcnA was extracted by alkaline lysis</p> | ||
| - | <p><strong>Gels</strong></p> | + | <p><strong><u>Gels</u></strong></p> |
<p>We run a 2% Agarose gel with the following samples:</p> | <p>We run a 2% Agarose gel with the following samples:</p> | ||
<ol> | <ol> | ||
| Line 437: | Line 438: | ||
</ol> | </ol> | ||
<p>With this gel the parts were verified</p> | <p>With this gel the parts were verified</p> | ||
| - | <p><strong>PCR</strong></p> | + | <p><strong><u>PCR </u></strong></p> |
<p>We performed a PCR reaction for RcnA and part 1 using Taq pol.</p> | <p>We performed a PCR reaction for RcnA and part 1 using Taq pol.</p> | ||
| - | <p><strong> | + | <p><strong><u>Transformation y ligation</u></strong></p> |
<p>We took cut RcnA and then it was ligated at vector PBBIMCS_5</p> | <p>We took cut RcnA and then it was ligated at vector PBBIMCS_5</p> | ||
<table width="30%" border="1" cellpadding="0" cellspacing="0"> | <table width="30%" border="1" cellpadding="0" cellspacing="0"> | ||
| Line 468: | Line 469: | ||
</table> | </table> | ||
<p>The transformation was performed by the previously mentioned technique and the strain was cultured in two Gentamycine cages(Gm 20)</p> | <p>The transformation was performed by the previously mentioned technique and the strain was cultured in two Gentamycine cages(Gm 20)</p> | ||
| - | <p><strong>PCR Cleaning</strong></p> | + | <p><strong><u>PCR Cleaning </u></strong></p> |
<p>We cleaned the PCR and it was filled up to 40 μl</p> | <p>We cleaned the PCR and it was filled up to 40 μl</p> | ||
| + | </div></td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | <tr> | ||
| + | <td class="subHeader" bgcolor="#99CC66" id="13">2008-08-13</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong></p> | ||
| + | <p>4μl of each sample were charged in the following order:<br /> | ||
| + | </p> | ||
| + | <ol> | ||
| + | <li>Molecular Marker</li> | ||
| + | <li> RcnA 1</li> | ||
| + | <li> RcnA 3</li> | ||
| + | <li> RcnA 4</li> | ||
| + | <li> RcnA 5</li> | ||
| + | <li> RcnA 6</li> | ||
| + | <li> Part 1_1</li> | ||
| + | <li> Part 1_3</li> | ||
| + | <li> Part 1_6</li> | ||
| + | <li> Part 1_7</li> | ||
| + | <li> Part 1_9</li> | ||
| + | </ol> | ||
| + | <p><Falta pegar gel segun liber...></p> | ||
| + | <p><u><strong>Part 1_1 restriction (Double digestion)</strong></u></p> | ||
| + | <table width="23%" border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>36 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>EcoR1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BamH1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p>After double digestion of part 1 we left massive ligation of part 1,2 and 3.</p> | ||
| + | <p>Ligation Recipe:</p> | ||
| + | <table width="23%" border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Part 1</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Part 2</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Part 3</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 5x</p></td> | ||
| + | <td width="50%"><p>4 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Ligase</p></td> | ||
| + | <td width="50%"><p>1 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Total </strong></p></td> | ||
| + | <td width="50%"><p>20 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p>Restriction of the other bioparts was performed</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="198"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>RcnA_4</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>6 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 2</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>30 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Xba1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>HindiIII</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="198"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>RcnA_6</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>11 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 2</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>25 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Xba1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>HindiIII</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="198"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Part 1_3</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p> </td> | ||
| + | <td width="50%"><p>6 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer EcoR1</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>30 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>EcoR1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BamH1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="198"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Part 1_6</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>6 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer EcoR1</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>30 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>EcoR1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BamH1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="198"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Part 1_9</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>1 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer EcoR1</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BSA</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>35 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>EcoR1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>BamH1</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table></div></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="subHeader" bgcolor="#99CC66" id="14">2008-08-14</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong></p> | ||
| + | <p><strong><u>GEL</u></strong></p> | ||
| + | <p>2% Agarose gel was run with the product of the massive ligation of the three parts</p> | ||
| + | <p>(we didn't obtain the desired product)</p> | ||
| + | <p><strong><u>PCR</u></strong></p> | ||
| + | <p>We pick up a PCR product with the folowing oligos:</p> | ||
| + | <blockquote> | ||
| + | <p>a) 1up and 2low<br /> | ||
| + | b) 2up and 2low<br /> | ||
| + | c) control</p> | ||
| + | </blockquote> | ||
| + | <p>The PCR reaction was prepared in the following way:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="204"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Reaction 1</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>10 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 3.3x</p></td> | ||
| + | <td width="50%"><p>6 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>dNTPs</p></td> | ||
| + | <td width="50%"><p>4 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Oligo up</p></td> | ||
| + | <td width="50%"><p>2.5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Oligo low</p></td> | ||
| + | <td width="50%"><p>2.5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Mg (Ac)2</p></td> | ||
| + | <td width="50%"><p>3 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>2 μl </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <table width="27%" border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tr> | ||
| + | <td width="50%"><p><strong>Reaction 2</strong></p></td> | ||
| + | <td width="50%"><p> </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>9 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 3.3x</p></td> | ||
| + | <td width="50%"><p>9 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>rTth</p></td> | ||
| + | <td width="50%"><p>2 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p><strong><u>GEL </u></strong></p> | ||
| + | <p>We run for 1 hr, an 2% Agarose Gel </p> | ||
| + | <ol> | ||
| + | <li>Molecular Marker (2.5 μl)</li> | ||
| + | <li> P1_P2 (oligo 1 up y 2 low) ligation with part 3 (normal) (5μl)</li> | ||
| + | <li> P2_P3 normal (oligo 2 up y 3 low) (5 μl)</li> | ||
| + | <li> Negative Control(oligo 1 up y 2 low) (5 μl)</li> | ||
| + | <li> P1_P2 (oligo 1 up y 2 low) ligation with part 3 (mutated) (5 μl)</li> | ||
| + | <li> P2_P3 mutated (oligo 2 up y 3 low) (5μl)</li> | ||
| + | <li> Negative Control (oligo 1 up y 2 low) (5μl)</li> | ||
| + | </ol> | ||
| + | <p>*The results suggest inspecific primer join.</p> | ||
| + | <p><u><strong>Cultures</strong></u></p> | ||
| + | <p>We left cultures of the RcnA + PBBRIMCS_5 ligation</p> | ||
| + | </div></td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td class="subHeader" bgcolor="#99CC66" id="15">2008-08-15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong></p> | ||
| + | <p><strong><u>Ligation repeat</u></strong></p> | ||
| + | <p>Performed in order to obtain at least part 1 and part 2 joined</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0" width="201"> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Part 1</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Part 2</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer 5x</p></td> | ||
| + | <td width="50%"><p>4 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>T4DNA Pol</p></td> | ||
| + | <td width="50%"><p>1 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Total</p></td> | ||
| + | <td width="50%"><p>20 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p><strong><u>Ligation PCR of P1_P2</u></strong></p> | ||
| + | <table width="26%" border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tr> | ||
| + | <td width="50%"><p>H2O</p></td> | ||
| + | <td width="50%"><p>33 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Buffer</p></td> | ||
| + | <td width="50%"><p>5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>dNTPs</p></td> | ||
| + | <td width="50%"><p>2.5 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Oligo 1 up</p></td> | ||
| + | <td width="50%"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Oligo 2 low</p></td> | ||
| + | <td width="50%"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Mg(Cl)2</p></td> | ||
| + | <td width="50%"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>DNA</p></td> | ||
| + | <td width="50%"><p>1 μl</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Taq</p></td> | ||
| + | <td width="50%"><table border="0" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="50%"><p>1 μl</p></td> | ||
| + | </tr> | ||
| + | </table></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="50%"><p>Total</p></td> | ||
| + | <td width="50%"><p>50 μl</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p>*The ligation and the PCR were done ___</p> | ||
| + | <p><strong><u>Plasmid Extraction<br /> | ||
| + | </u></strong></p> | ||
| + | <p>Plasmid PBBIMCS_5 was extracted from the cultures of the last day.</p> | ||
| + | <p><strong><u>PCR ligation Gel of part1_part2</u></strong></p> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/0/0b/Gel_15Ago08.png" alt="Gel_15Ago08" width="400" /></p> </div></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="subHeader" bgcolor="#99CC66" id="17">2008-08-17</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="bodyText"><div align="justify"><p><strong>WET LAB:</strong></p> | ||
| + | <p><strong><u>Ligation part1+PRK415</u></strong></p> | ||
| + | <p>The tube DR of PRK415 was used along with the following samples of part 1:</p> | ||
| + | <ul> | ||
| + | <li>DR Part1_9/pJET (130808) </li> | ||
| + | <li> DR Part1_6/pJET (130808) </li> | ||
| + | <li> DR Part1_3/pJET (130808) </li> | ||
| + | </ul> | ||
| + | <p>Recipe for each ligation tube</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>Vector </p></td> | ||
| + | <td width="132" valign="top"><p>4 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>Part 1 </p></td> | ||
| + | <td width="132" valign="top"><p>8 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>Buffer 5x </p></td> | ||
| + | <td width="132" valign="top"><p>4 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>Enzyme </p></td> | ||
| + | <td width="132" valign="top"><p>1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>H2O </p></td> | ||
| + | <td width="132" valign="top"><p>3 μl </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p><strong><u>Restriction</u></strong></p> | ||
| + | <p>Linearization of pBB to use it as a control in RcnA+pBB Gel</p> | ||
| + | <p>Recipe for each restriction tube:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>H20 </p></td> | ||
| + | <td width="132" valign="top"><p>10 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>Buffer </p></td> | ||
| + | <td width="132" valign="top"><p>3 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>DNA </p></td> | ||
| + | <td width="132" valign="top"><p>16 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p>EcoR1 </p></td> | ||
| + | <td width="132" valign="top"><p>1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="79" valign="top"><p> </p></td> | ||
| + | <td width="132" valign="top"><p><strong>30 μl </strong></p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p><strong><u>Plasmid Extraction Gel pBB+RcnA</u></strong></p> | ||
| + | <ol> | ||
| + | <li>Molecular Marker (2 μl) </li> | ||
| + | <li> [1] Cage2 /11_1 </li> | ||
| + | <li> [2] Cage1 /1_2 </li> | ||
| + | <li> [3] Cage2B /1_1 </li> | ||
| + | <li> [4] Cage2a /10_1 </li> | ||
| + | <li> [5] Cage2b /8_1 </li> | ||
| + | <li> [6] Cage2b /6_1 </li> | ||
| + | <li> [7] Cage1 /5_1 </li> | ||
| + | <li> [8] Cage1 /2_1 </li> | ||
| + | <li> [9] Cage2b /4_1</li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/f/ff/Gel_17Ago08.png" alt="Gel_17Ago08" width="400" /></p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p> </p></td> | ||
| + | <td width="82" valign="top"><p>1 </p></td> | ||
| + | <td width="82" valign="top"><p>2 </p></td> | ||
| + | <td width="82" valign="top"><p>3 </p></td> | ||
| + | <td width="82" valign="top"><p>4 </p></td> | ||
| + | <td width="82" valign="top"><p>5 </p></td> | ||
| + | <td width="82" valign="top"><p>6 </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p>Water </p></td> | ||
| + | <td width="82" valign="top"><p>7 μl </p></td> | ||
| + | <td width="82" valign="top"><p>7 μl </p></td> | ||
| + | <td width="82" valign="top"><p>7 μl </p></td> | ||
| + | <td width="82" valign="top"><p>5 μl </p></td> | ||
| + | <td width="82" valign="top"><p>7 μl </p></td> | ||
| + | <td width="82" valign="top"><p>7 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p>Buffer </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | <td width="82" valign="top"><p>2 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p>DNA </p></td> | ||
| + | <td width="82" valign="top"><p>10 μl </p></td> | ||
| + | <td width="82" valign="top"><p>10 μl </p></td> | ||
| + | <td width="82" valign="top"><p>10 μl </p></td> | ||
| + | <td width="82" valign="top"><p>12 μl </p></td> | ||
| + | <td width="82" valign="top"><p>10 μl </p></td> | ||
| + | <td width="82" valign="top"><p>10 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p>Enzyme </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | <td width="82" valign="top"><p>1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="82" valign="top"><p> </p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | <td width="82" valign="top"><p><strong>20 μl </strong></p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <p>*In order to verify the size of the insert in pBB</p> | ||
| + | <p><strong><u>Extraction plasmid restrictions (pBB+RcnA) with EcoR1</u></strong></p> | ||
| + | <p>We used:</p> | ||
| + | <ul> | ||
| + | <li>[1] Cage2 /11_1 </li> | ||
| + | <li> [2] Cage1 /1_2 </li> | ||
| + | <li> [3] Cage2B /1_1 </li> | ||
| + | <li> [4] Cage2a /10_1 </li> | ||
| + | <li> [5] Cage2b /8_1 </li> | ||
| + | <li> [9] Cage2b /4_1 </li> | ||
| + | </ul> | ||
| + | <p><strong><u>Gel </u></strong></p> | ||
| + | <ol> | ||
| + | <li>Molecular Marker (2.5 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 2b/4 (2 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 2a/10 (2 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 2b/5 (2 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 2a/14(2 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 2a/9(2 μl) </li> | ||
| + | <li> Restriction EcoR1 pBBIMCS-5 limpio con kit(2 μl) </li> | ||
| + | <li> Restriction EcoR1 BamH1 PRK415 R1(2 μl) </li> | ||
| + | <li> Restriction EcoR1 BamH1 PRK415 R2(2 μl </li> | ||
| + | <li> PCR control1 biopart 1 oligo 1up 2low (5 μl) </li> | ||
| + | <li> PCR control1 biopart 2 oligo 1up 2low (5 μl) </li> | ||
| + | <li> Negative Control PCR (5 μl) </li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/0/0a/Gel_16Ago08.png" alt="Gel_16Ago08" width="400"/></p> | ||
| + | <p>Conclusions of the previous gel:</p> | ||
| + | <ul> | ||
| + | <li>The Oligos 1up and 2low don't join in unexpected sites</li> | ||
| + | <li>RcnA wasn't inserted in pBB</li> | ||
| + | <li>It's necessary to repeat the double restriction of PRK</li> | ||
| + | </ul> | ||
| + | <p><strong><u>PCR</u> </strong></p> | ||
| + | <ol> | ||
| + | <li>Double ligation</li> | ||
| + | <li> Control without DNA </li> | ||
| + | <li> Control without oligos (¿Is there something contaminated?) </li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/2/20/Gel_16Ago08_2.png" alt="Gel_16Ago08_2" width="400"/></p> | ||
| + | <p>We run a gel of this three PCRs and we didn't obtain any product.</p> | ||
| + | <p><strong><u>Repetición PCR</u> </strong></p> | ||
| + | <p>We repeated the previous PCR reaction using the following recipe:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>DNA </p></td> | ||
| + | <td width="120" valign="top"><p>2 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>H2O </p></td> | ||
| + | <td width="120" valign="top"><p>32 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>dNTP’s </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Mg(Cl)2 </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Oligo 1up </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Oligo 2low </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Taq </p></td> | ||
| + | <td width="120" valign="top"><p>1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Buffer </p></td> | ||
| + | <td width="120" valign="top"><p>5 μl </p></td> | ||
| + | </tr> | ||
| + | </table></div></td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td class="subHeader" bgcolor="#99CC66" id="19">2008-08-19</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td class="bodyText"><div align="justify"><p><strong>WET LAB</strong></p> | ||
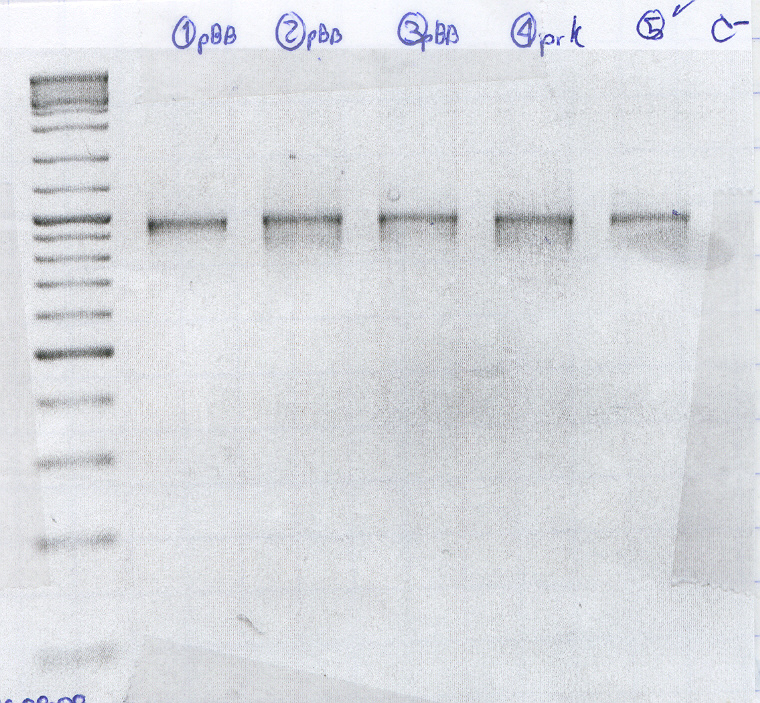
| + | <p><u><strong>Restrictions gel pBB+RcnA</strong></u></p> | ||
| + | <ol> | ||
| + | <li>Molecular Marker (2.5 μl) </li> | ||
| + | <li> [1] Cage2 /11_1 </li> | ||
| + | <li> [2] Cage1 /1_2 </li> | ||
| + | <li> [3] Cage2B /1_1 </li> | ||
| + | <li> [4] Cage2a /10_1 </li> | ||
| + | <li> [5] Cage2b /8_1 </li> | ||
| + | <li> [9] Cage2b /4_ </li> | ||
| + | <li> pBB1 </li> | ||
| + | <li> pBB2 </li> | ||
| + | <li> pBB3<br /> | ||
| + | </li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/e/e7/Gel_19_Ago_08.png" alt="Gel_19Ago08" width="400" /></p> | ||
| + | <p>Cultured from a strain result of the transformation</p> | ||
</div></td> | </div></td> | ||
</tr> | </tr> | ||
| Line 493: | Line 1,125: | ||
<p> | <p> | ||
<br /> | <br /> | ||
| - | <a name="20ago"></a><b | + | <a name="20ago"></a><b>MODELING:</b><br> |
Reaction 3, AHL:LuxR<br /> | Reaction 3, AHL:LuxR<br /> | ||
Conflict: k3 (ON) <k3 (OFF)? <br /> | Conflict: k3 (ON) <k3 (OFF)? <br /> | ||
| Line 508: | Line 1,140: | ||
This explains the criteria they used to determine their parameters: <br /> | This explains the criteria they used to determine their parameters: <br /> | ||
<<For each layout we attempted to identify a set of parameters that optimize the functional fitness of the network. The search in the parameter space is constrained by requesting that the kinetic parameters must remain in the biologically realistic range and the resulting network should demonstrate the behavior compatible with our present understanding of the phenomenon quorum sensing.>></p> | <<For each layout we attempted to identify a set of parameters that optimize the functional fitness of the network. The search in the parameter space is constrained by requesting that the kinetic parameters must remain in the biologically realistic range and the resulting network should demonstrate the behavior compatible with our present understanding of the phenomenon quorum sensing.>></p> | ||
| + | <p><strong>WET LAB</strong></p> | ||
| + | <p><strong><u id="">Agarose Gel 1% low fusion point for band purification</u></strong></p> | ||
| + | <ol> | ||
| + | <li>Gel1</li> | ||
| + | <li> Molecular Weight marker</li> | ||
| + | <li> [1] </li> | ||
| + | <li> [2] </li> | ||
| + | <li>[3] </li> | ||
| + | </ol> | ||
| + | <p><u><strong><br /> | ||
| + | Ligation PCR parte1+parte2(35 ciclos)</strong></u></p> | ||
| + | <ol> | ||
| + | <li> 1_2 oligo 1up-2lower </li> | ||
| + | <li> 1_1 oligo 1up-1lower </li> | ||
| + | <li> 2_2 oligo 2up-2lower </li> | ||
| + | <li> 1_3 oligo 1up-3lower </li> | ||
| + | <li> 1_2 oligo 1up-2lower</li> | ||
| + | </ol> | ||
| + | <p>Recipes:</p> | ||
| + | <table border="1" cellspacing="0" cellpadding="0"> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>DNA </p></td> | ||
| + | <td width="120" valign="top"><p>2 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Water</p></td> | ||
| + | <td width="120" valign="top"><p>32 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>dNTP’s </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Mg(Cl)2 </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Oligo 1up </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Oligo 2low </p></td> | ||
| + | <td width="120" valign="top"><p>2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Taq </p></td> | ||
| + | <td width="120" valign="top"><p>1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="91" valign="top"><p>Buffer </p></td> | ||
| + | <td width="120" valign="top"><p>5 μl </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
</div></td> | </div></td> | ||
</tr> | </tr> | ||
| Line 516: | Line 1,202: | ||
<td class="bodyText"><div align="justify"><p> | <td class="bodyText"><div align="justify"><p> | ||
<strong> | <strong> | ||
| - | + | MODELING:</strong><br /> | |
<br /> | <br /> | ||
Reaction 4: Natural degradation of cI*<br /> | Reaction 4: Natural degradation of cI*<br /> | ||
| Line 565: | Line 1,251: | ||
Peter<br /> | Peter<br /> | ||
<br /> | <br /> | ||
| - | |||
| - | |||
</p> | </p> | ||
| + | <p><strong>WET LAB</strong></p> | ||
| + | <p><strong><u>Agarose Gel 1% low fusion point for band purification</u></strong></p> | ||
| + | <ol> | ||
| + | <li>Molecular Weight Marker </li> | ||
| + | <li> [1] Cage2 /11_1 pBB+RcnA </li> | ||
| + | <li> [2] Cage1 /1_2 pBB+RcnA </li> | ||
| + | <li> [5] Cage2b /8_1 pBB+RcnA </li> | ||
| + | <li> [9] Cage2b /4_1 pBB+RcnA <br /> | ||
| + | </li> | ||
| + | </ol> | ||
| + | <p><strong><u>Gel Purification PCR of pBB+rcnA</u> </strong></p> | ||
| + | <ol> | ||
| + | <li> Purification 1</li> | ||
| + | <li> Purification 2 </li> | ||
| + | <li> Purification 3</li> | ||
| + | <li> Purification 4 clean PRK</li> | ||
| + | <li> Positive control</li> | ||
| + | <li>Negative control</li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/8/84/Gel_21Ago08.png" alt="Gel28Aug08" width="400" /></p> | ||
| + | <p>The cells seem to carry the plasmid with RcnA, However one of the negative controls gave product.</p> | ||
| + | <p><br /> | ||
| + | All of them were positive, however there are some spurious products.(no photo was added because the low melting gels are quite fragile and we couldn't carry it to the transiluminator with camera.)</p> | ||
| + | <p> Recipe: </p> | ||
| + | <table border="1" cellpadding="0" cellspacing="0"> | ||
| + | <tbody > | ||
| + | <tr > | ||
| + | <td valign="top" width="91"><p> DNA </p></td> | ||
| + | <td valign="top" width="120"><p> 2 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="91"><p> Water </p></td> | ||
| + | <td valign="top" width="120"><p> 32 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top" width="91"><p> dNTP’s </p></td> | ||
| + | <td valign="top" width="120"><p > 2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td id="" valign="top" width="91"><p id=""> Mg(Cl)2 </p></td> | ||
| + | <td id="" valign="top" width="120"><p id=""> 2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr id=""> | ||
| + | <td id="" valign="top" width="91"><p id=""> Oligo 1up </p></td> | ||
| + | <td id="" valign="top" width="120"><p id=""> 2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr id=""> | ||
| + | <td id="" valign="top" width="91"><p id=""> Oligo 2low </p></td> | ||
| + | <td id="" valign="top" width="120"><p id=""> 2.5 μl </p></td> | ||
| + | </tr> | ||
| + | <tr id=""> | ||
| + | <td id="" valign="top" width="91"><p id=""> Taq </p></td> | ||
| + | <td id="" valign="top" width="120"><p id=""> 1 μl </p></td> | ||
| + | </tr> | ||
| + | <tr id=""> | ||
| + | <td id="" valign="top" width="91"><p id=""> Buffer </p></td> | ||
| + | <td id="" valign="top" width="120"><p id=""> 5 μl </p></td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <p id=""> A gel with the extraction of the plasmid PRK415+part_1 was run.</p> | ||
| + | <p><u><strong>Gel Electrophoresis of PCR_Ligation part1+part2 </strong></u></p> | ||
| + | <ol> | ||
| + | <li>[5] 1_2 oligo 1up-2lower</li> | ||
| + | <li>[4] 1_2 oligo 1up-2lower</li> | ||
| + | <li>[3] 1_2 oligo 1up-2lower</li> | ||
| + | <li>Molecular marker</li> | ||
| + | <li>[2] 1_2 oligo 1up-2lower</li> | ||
| + | <li>[1] 1_2 oligo 1up-2lower </li> | ||
| + | </ol> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2008/3/37/Gel_21Ago08_2.png" alt="Gel_21Ago08_2" width="400" /></p> | ||
| + | <p>We have got part1+part2 ligation</p> | ||
</div></td> | </div></td> | ||
</tr> | </tr> | ||
| Line 575: | Line 1,331: | ||
<tr> | <tr> | ||
<td class="bodyText"><div align="justify"><p> | <td class="bodyText"><div align="justify"><p> | ||
| - | <b | + | <b>MODELING:</b><br> |
Checking parameters:<br /> | Checking parameters:<br /> | ||
<br /> | <br /> | ||
| Line 590: | Line 1,346: | ||
<tr> | <tr> | ||
<td class="bodyText"><div align="justify"><p> | <td class="bodyText"><div align="justify"><p> | ||
| - | <strong | + | <strong>MODELING: </strong><br /> |
<br> | <br> | ||
To-do List | To-do List | ||
Revision as of 01:57, 29 October 2008
 |
LCG-UNAM-Mexico |  |
|||||||||||||
| iGEM 2008 TEAM | |||||||||||||||
|
|||||||||||||||
 "
"