Team:Cambridge/Signalling/Vectors
From 2008.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Cambridge/Notitle}} | {{Cambridge/Notitle}} | ||
| - | |||
{{Cambridge08}} | {{Cambridge08}} | ||
{{Cambridge08signalling}} | {{Cambridge08signalling}} | ||
<html> | <html> | ||
| - | < | + | <div style="background:#444; padding-right:15px; padding-left:15px;"> |
| - | + | ||
<b class="b1f"></b><b class="b2f"></b><b class="b3f"></b><b class="b4f"></b> | <b class="b1f"></b><b class="b2f"></b><b class="b3f"></b><b class="b4f"></b> | ||
<div class="contentf"> | <div class="contentf"> | ||
| - | <div style="background:#ddd; text-align:left | + | <div style="background:#ddd; text-align:left;"> |
</html> | </html> | ||
| Line 20: | Line 18: | ||
The integration site of these is pre-determined | The integration site of these is pre-determined | ||
| - | {| class="wikitable" border="1 | + | {| class="wikitable" border="1" |
|- | |- | ||
! BGSC Accession | ! BGSC Accession | ||
| Line 136: | Line 134: | ||
The user inserts a homologous piece of the Bacillus chromosome and the vector integrates there. | The user inserts a homologous piece of the Bacillus chromosome and the vector integrates there. | ||
| - | {| class="wikitable" border="1 | + | {| class="wikitable" border="1" |
|- | |- | ||
! BGSC Accession | ! BGSC Accession | ||
| Line 195: | Line 193: | ||
= Shuttle Vectors = | = Shuttle Vectors = | ||
| - | {| class="wikitable" border="1 | + | {| class="wikitable" border="1" |
|- | |- | ||
! BGSC Accession | ! BGSC Accession | ||
| Line 232: | Line 230: | ||
| [[image:ECE188.jpg|thumb|ECE188 Map]] | | [[image:ECE188.jpg|thumb|ECE188 Map]] | ||
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC02.htm From Bayreuth] | | [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC02.htm From Bayreuth] | ||
| - | | fontcolor="white"| Not yet checked, DNA stocks | + | | bgcolor="white" fontcolor="white"| Not yet checked, DNA stocks |
|- | |- | ||
| ECE189 | | ECE189 | ||
| Line 242: | Line 240: | ||
| [[image:ECE188.jpg|thumb|ECE189 Map]] | | [[image:ECE188.jpg|thumb|ECE189 Map]] | ||
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC04.htm From Bayreuth] | | [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC04.htm From Bayreuth] | ||
| - | | fontcolor="white"| Not yet checked, DNA stocks | + | | bgcolor="white" fontcolor="white"| Not yet checked, DNA stocks |
|- | |- | ||
| ECE190 | | ECE190 | ||
| Line 258: | Line 256: | ||
== [[IGEM:Cambridge/2008/Notebook/Turing_Pattern_Formation/2008/08/04|Plasmid Gels]] == | == [[IGEM:Cambridge/2008/Notebook/Turing_Pattern_Formation/2008/08/04|Plasmid Gels]] == | ||
* We ran these plasmids on gels, using double and single digests, to check them | * We ran these plasmids on gels, using double and single digests, to check them | ||
| + | <html> | ||
| + | </div></div> | ||
| + | <b class="b4f"></b><b class="b3f"></b><b class="b2f"></b><b class="b1f"></b> | ||
| + | </td></tr></table></div> | ||
| + | </html> | ||
Revision as of 04:35, 29 October 2008
All these vectors were kindly provided by Dr. Dan Ziegler, Director of the Bacillus Genetic Stock Center at Ohio State University.
Contents |
Integration Vectors
These vectors integrate into the chromosome and do not have a replication origin in Bacillus. They either integrate cassettes that require double crossovers and two homologous regions, or whole-plasmid insertions that only require one crossover and one corresponding region of homology.
Ectopic Integration
The integration site of these is pre-determined
| BGSC Accession | Vector Name | Features | Integration Locus | Vector Map | Vector Sequence | Possible Uses | Necessity (1-5 stars) | Checked on Gel |
|---|---|---|---|---|---|---|---|---|
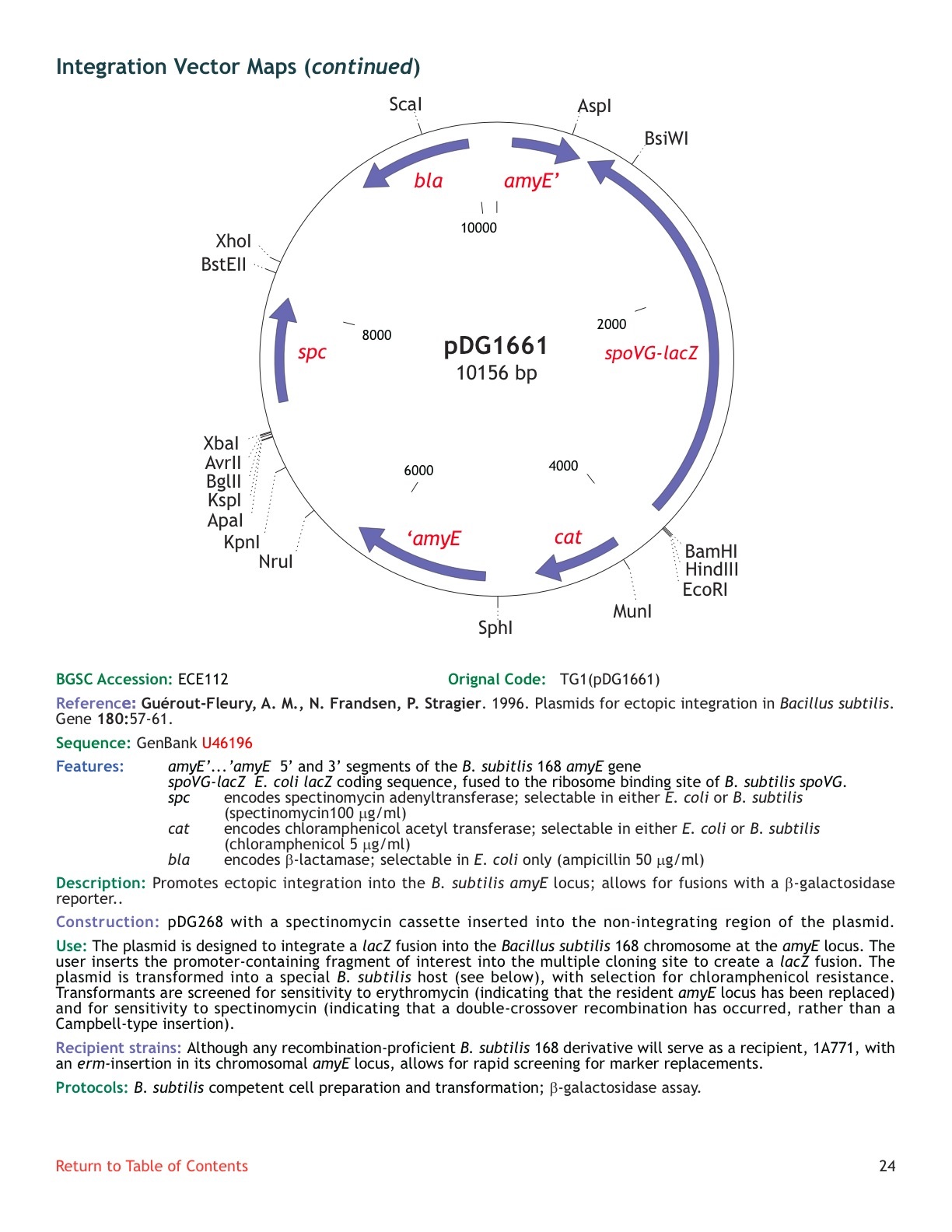
| ECE112 | pDG1661 |
| amyE | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=1185558 U46196] |
| **** | Yes | |
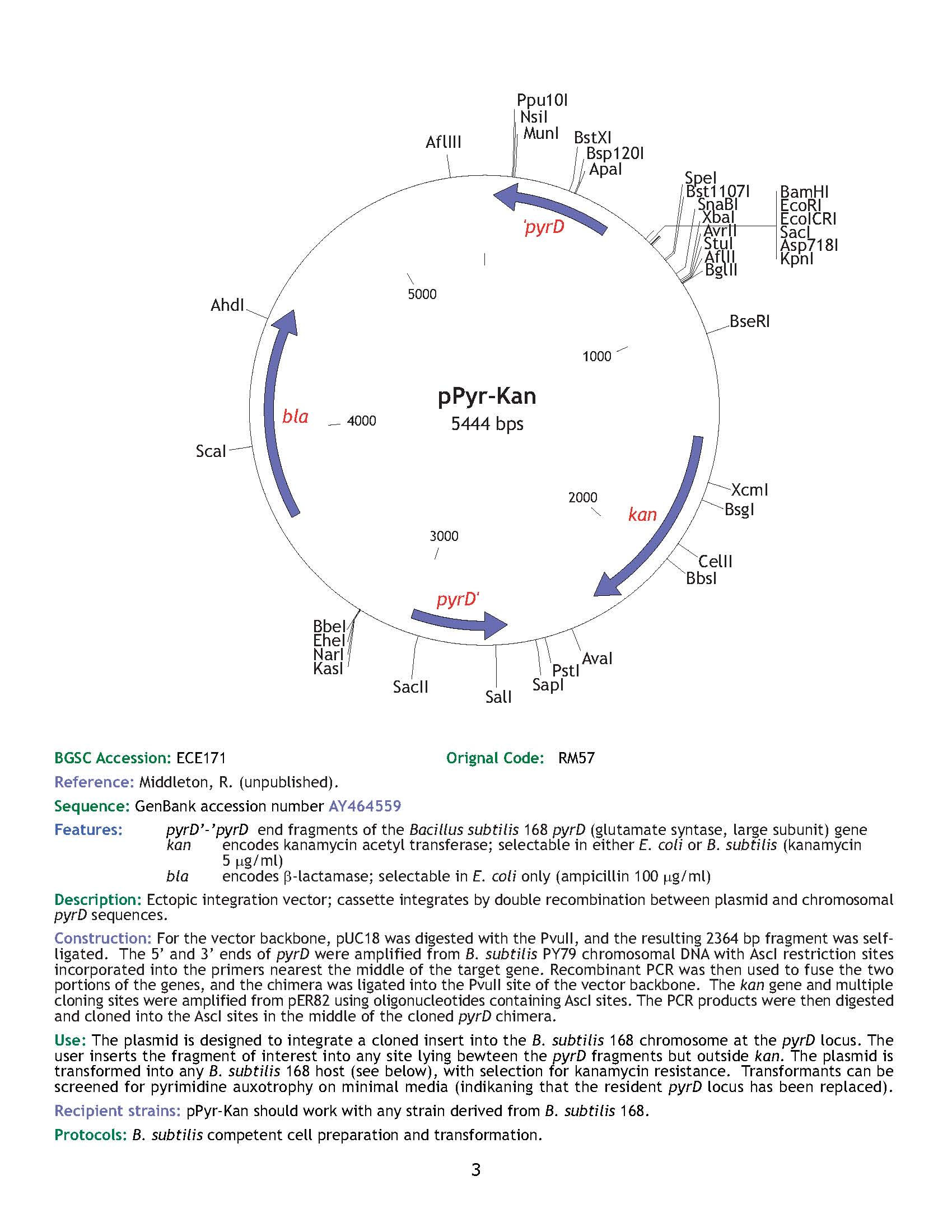
| ECE171 | pPyr-Kan |
| pyrD | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=39726213 AY464559] |
| **** | Yes | |
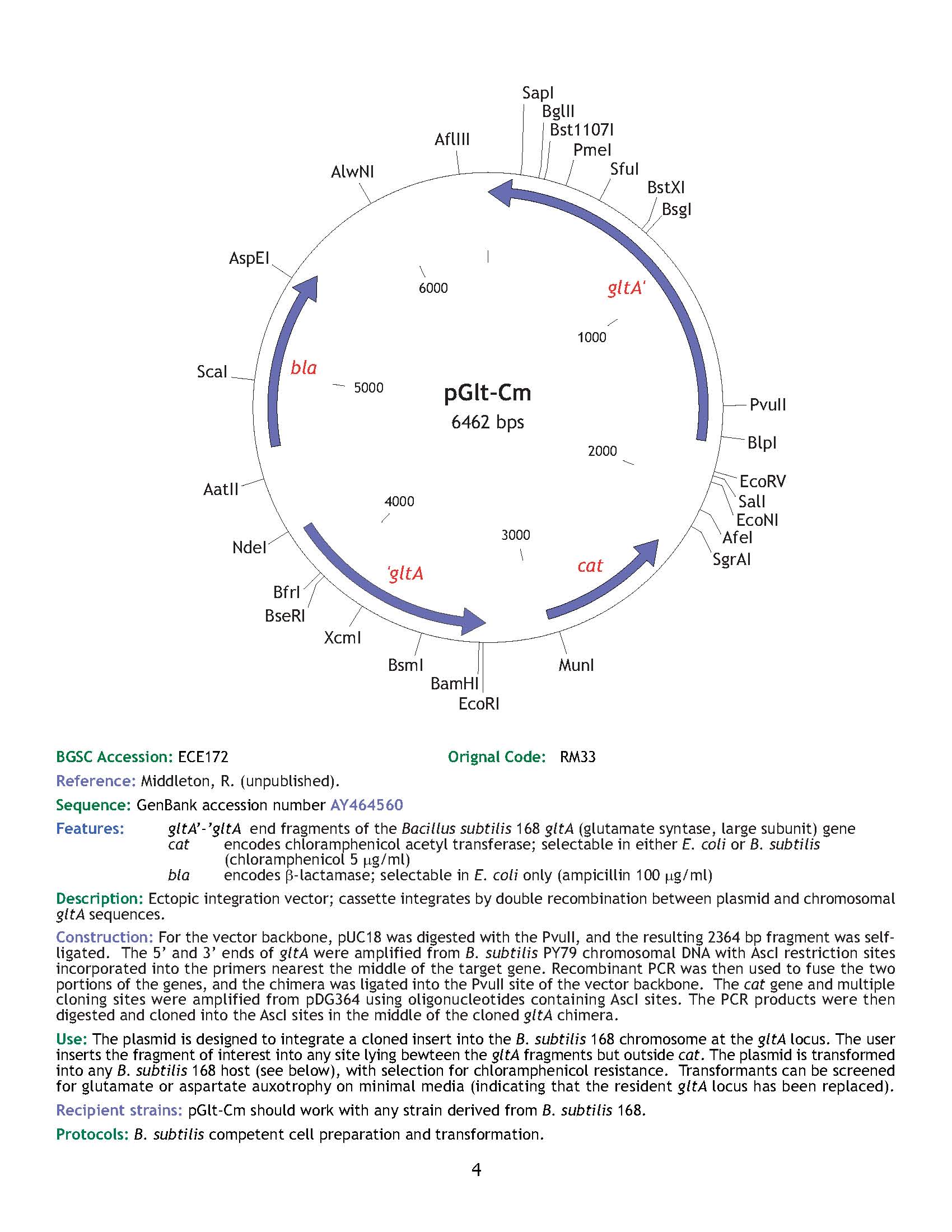
| ECE172 | pGlt-Cm |
| gltA | [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=39726216 AY464560] |
| *: can use PyrD & AmyE instead | No: wrong size | |
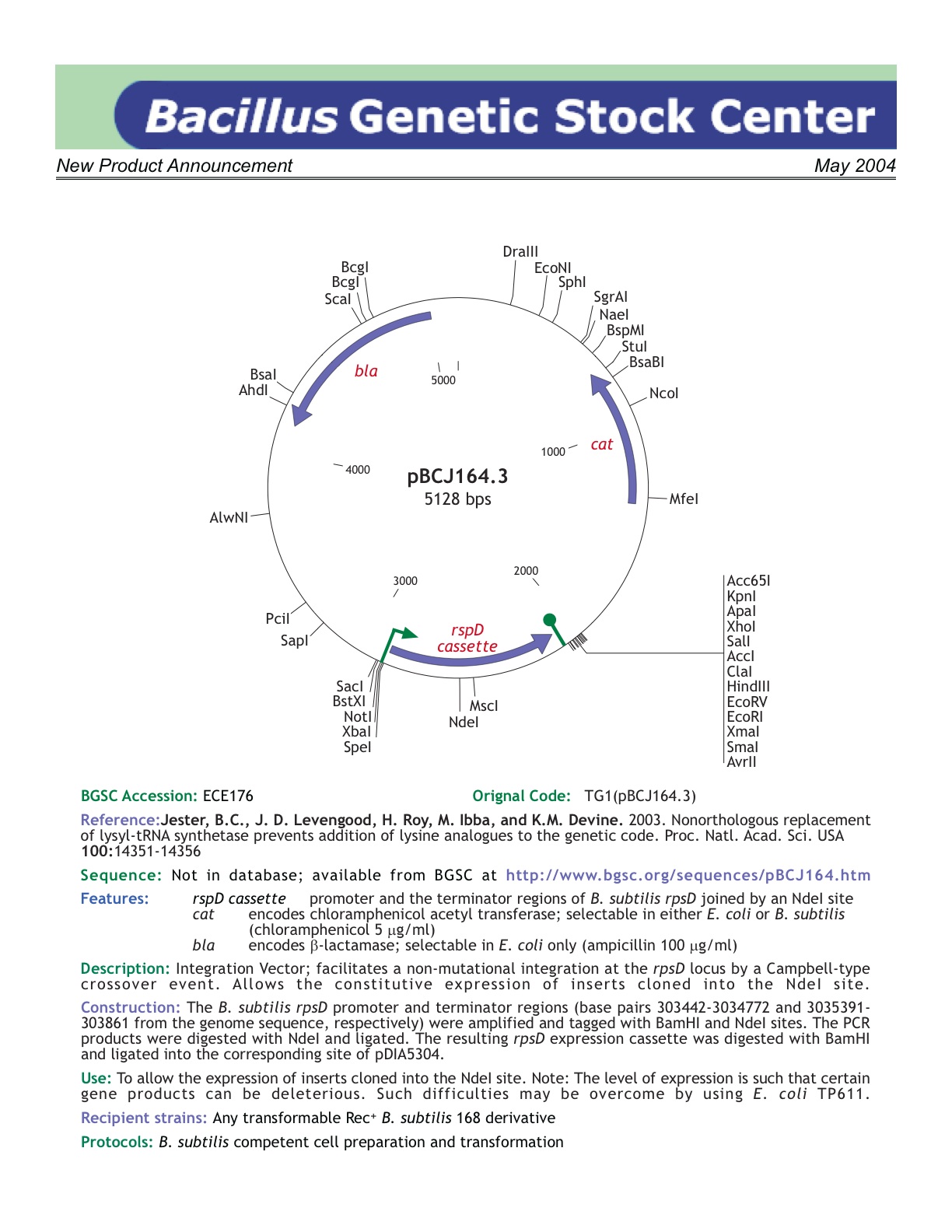
| ECE176 | pBCJ164 |
| rspD | [http://www.bgsc.org/sequences/pBCJ164.htm BGSC sequence] |
| *: use of strong rpsD promoter, but hard to insert into | Yes | |
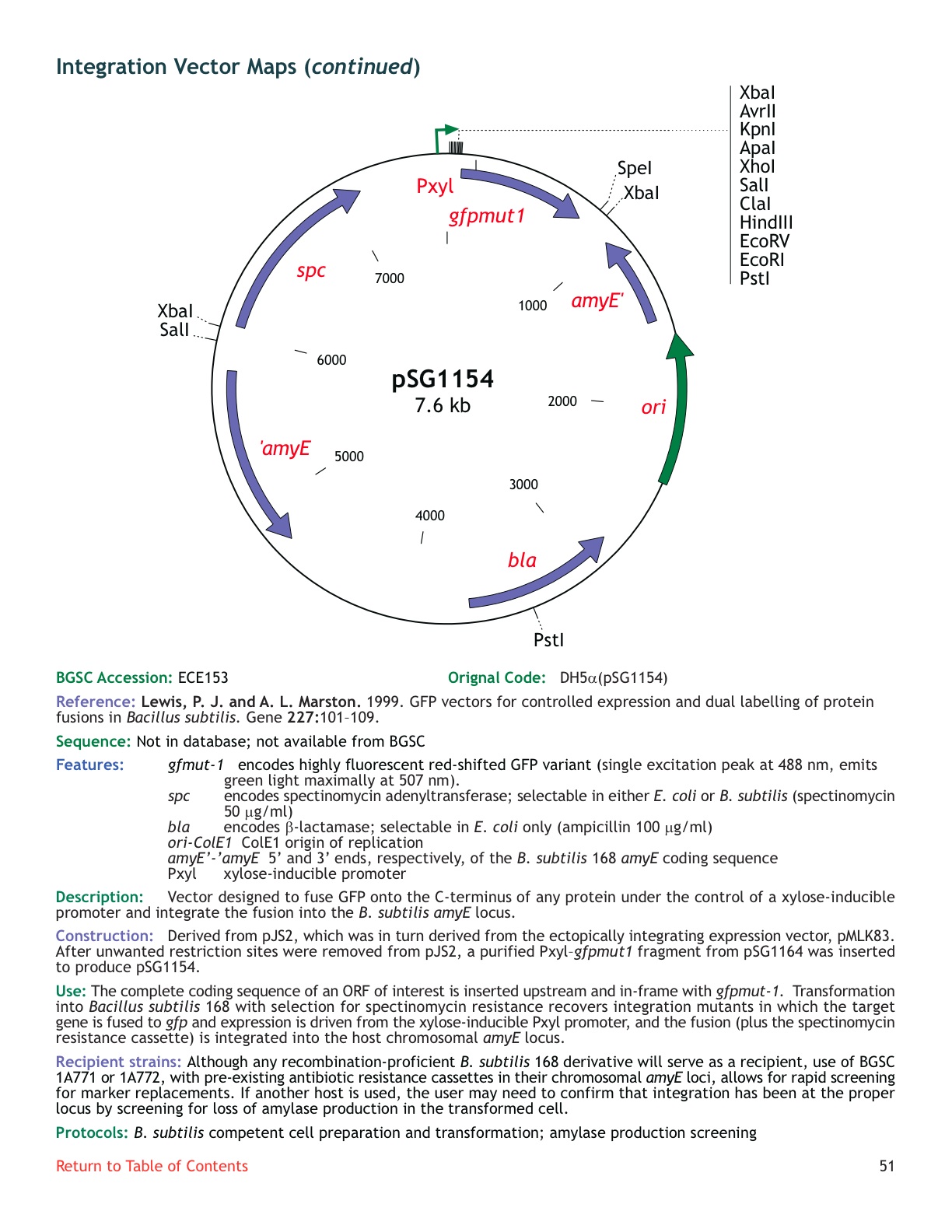
| ECE153 | pSG1154 |
| amyE | not available |
| ***: inducible promoter, strong gfp, no sequence | yes | |
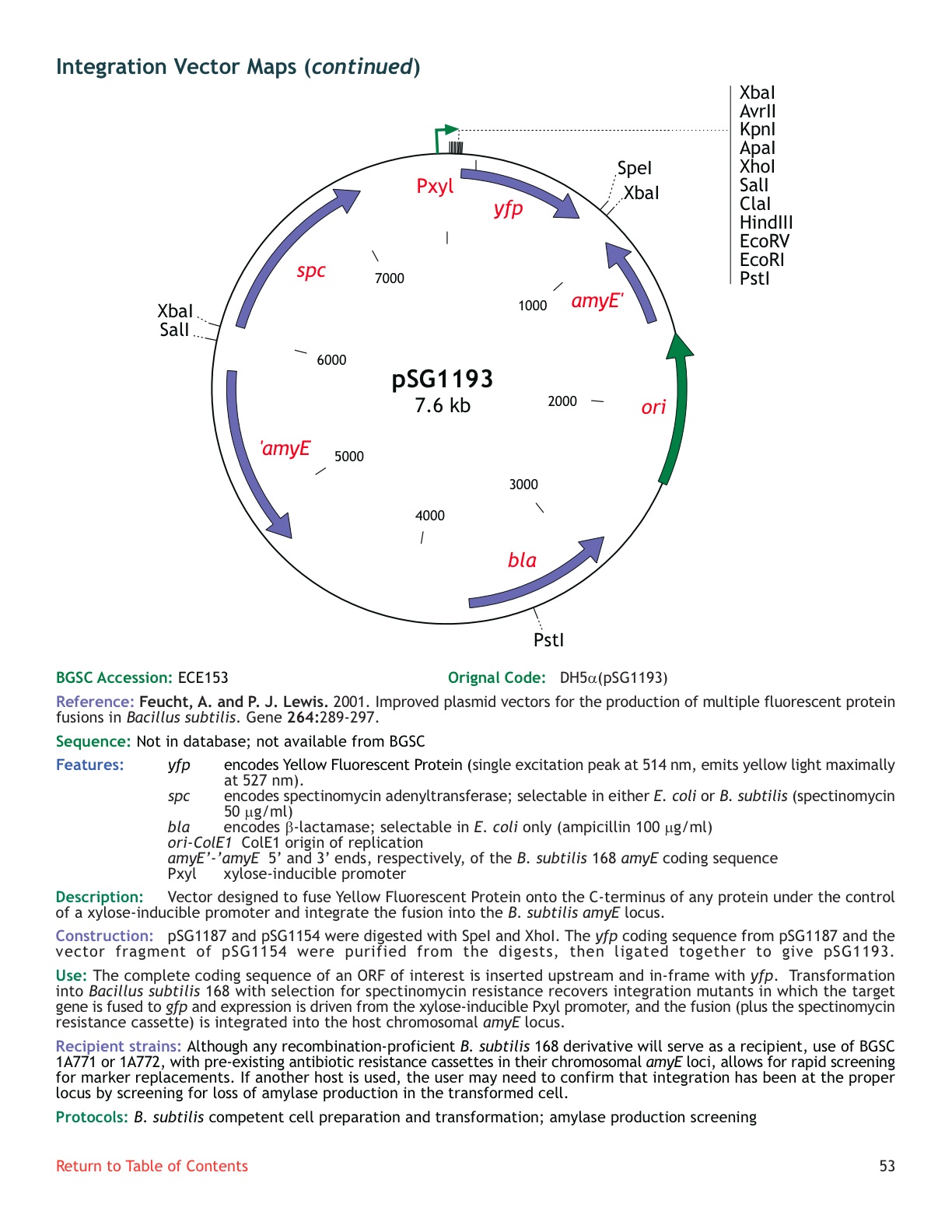
| ECE162 | pSG1193 |
| amyE | not available |
| ***: inducible promoter, strong gfp, no sequence | inconclusive : only one band |
Insert Integration
The user inserts a homologous piece of the Bacillus chromosome and the vector integrates there.
| BGSC Accession | Vector Name | Features | Vector Map | Vector Sequence | Checked on gel |
|---|---|---|---|---|---|
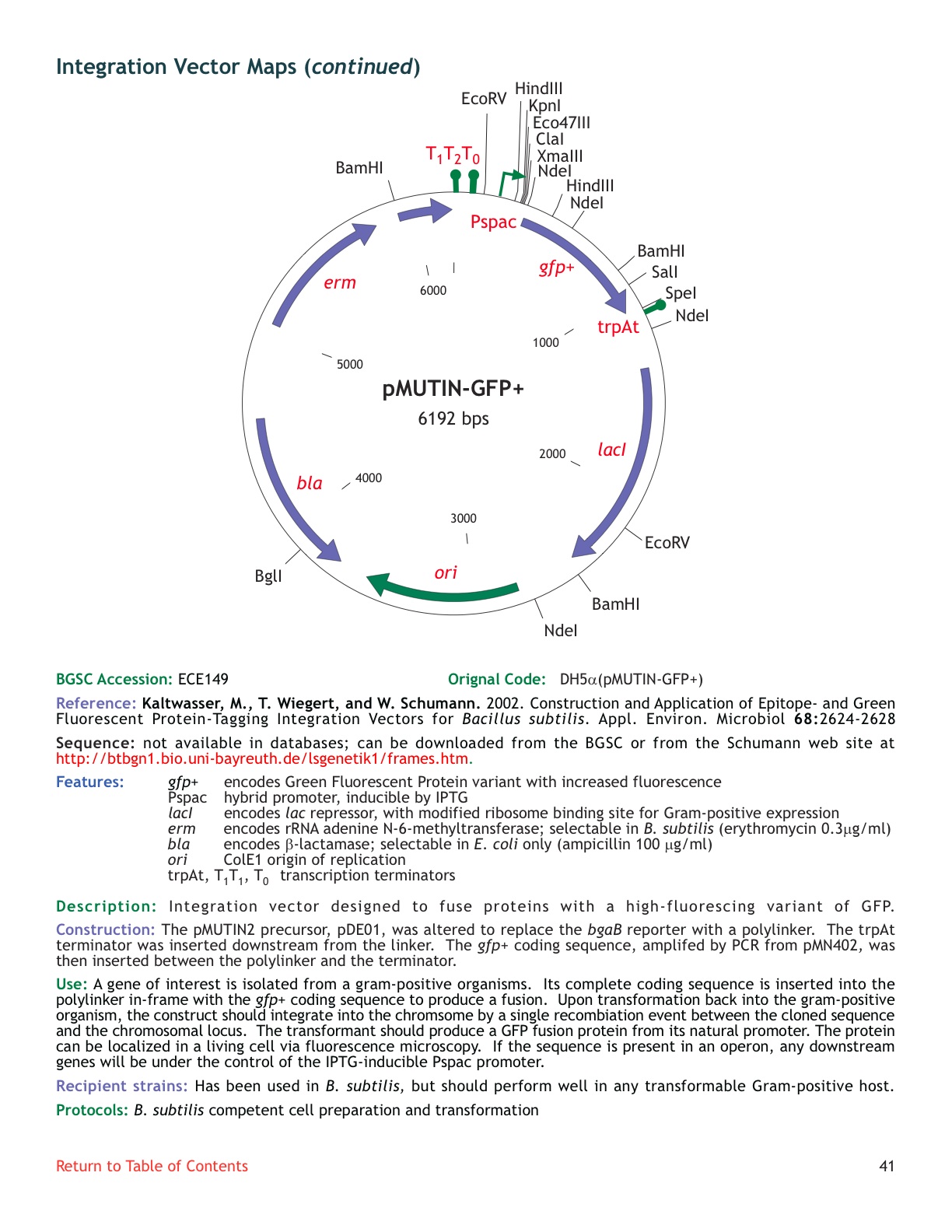
| ECE149 | pMUTIN-GFP+ |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pmutingfp.htm From Bayreuth] | No: 3 bands on the gel | |
| ECE150 | pMUTIN-CFP |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pmutincfp.htm From Bayreuth] (sequence incomplete) | yes | |
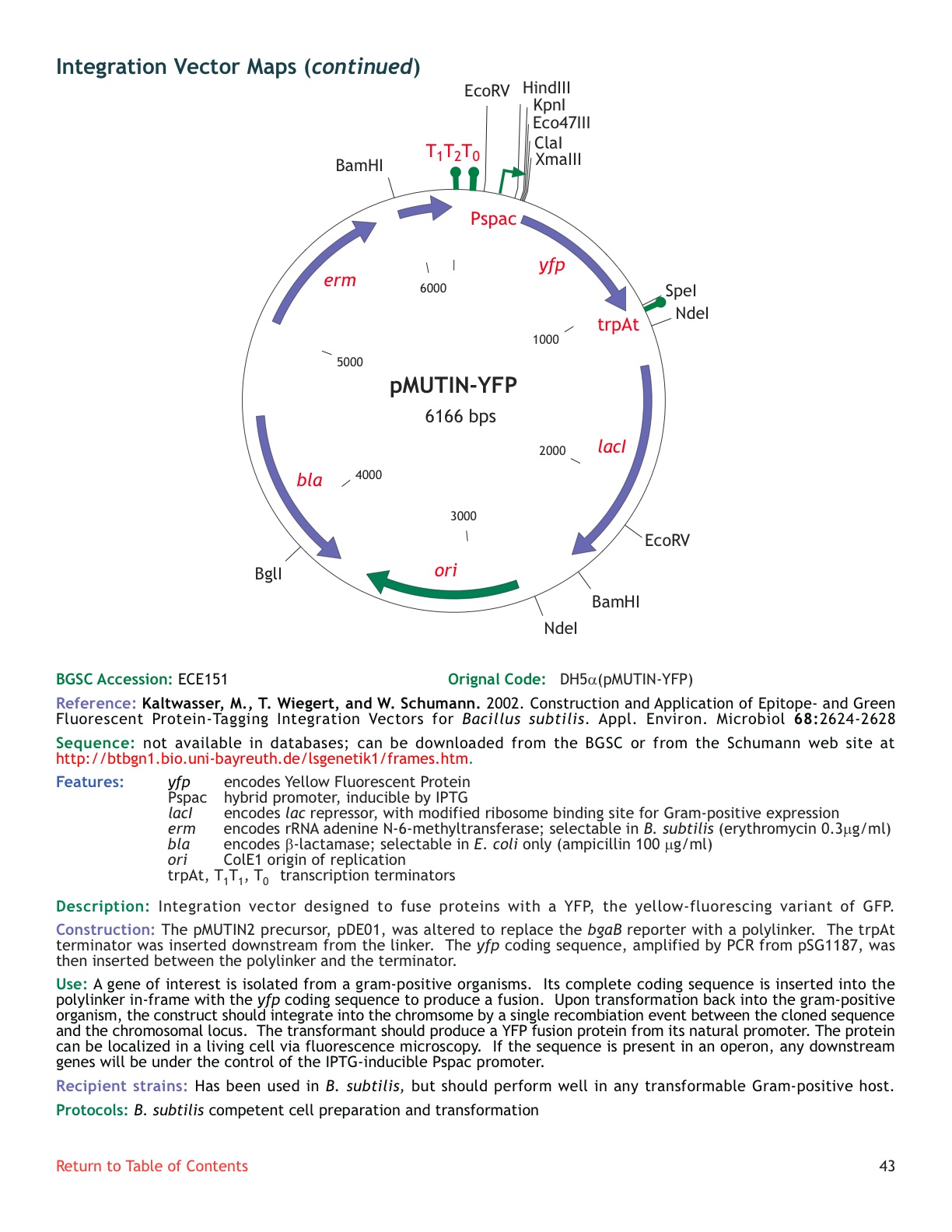
| ECE151 | pMUTIN-YFP |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pmutinyfp.htm From Bayreuth] | Yes | |
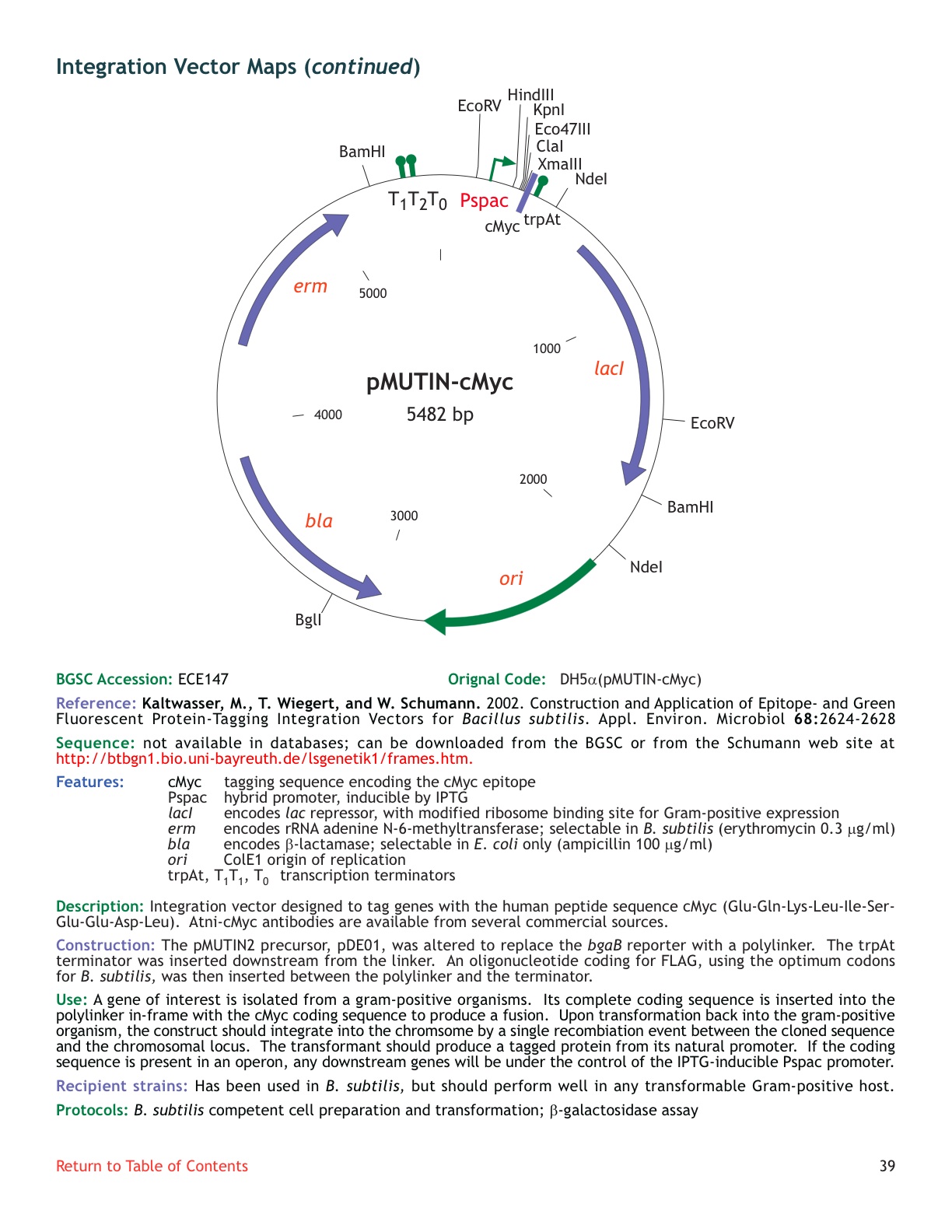
| ECE147 | pMUTIN-cMyc |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pmutincmyc.htm From Bayreuth] | yes |
Shuttle Vectors
| BGSC Accession | Vector Name | Features | Vector Map | Vector Sequence | Checked on gel |
|---|---|---|---|---|---|
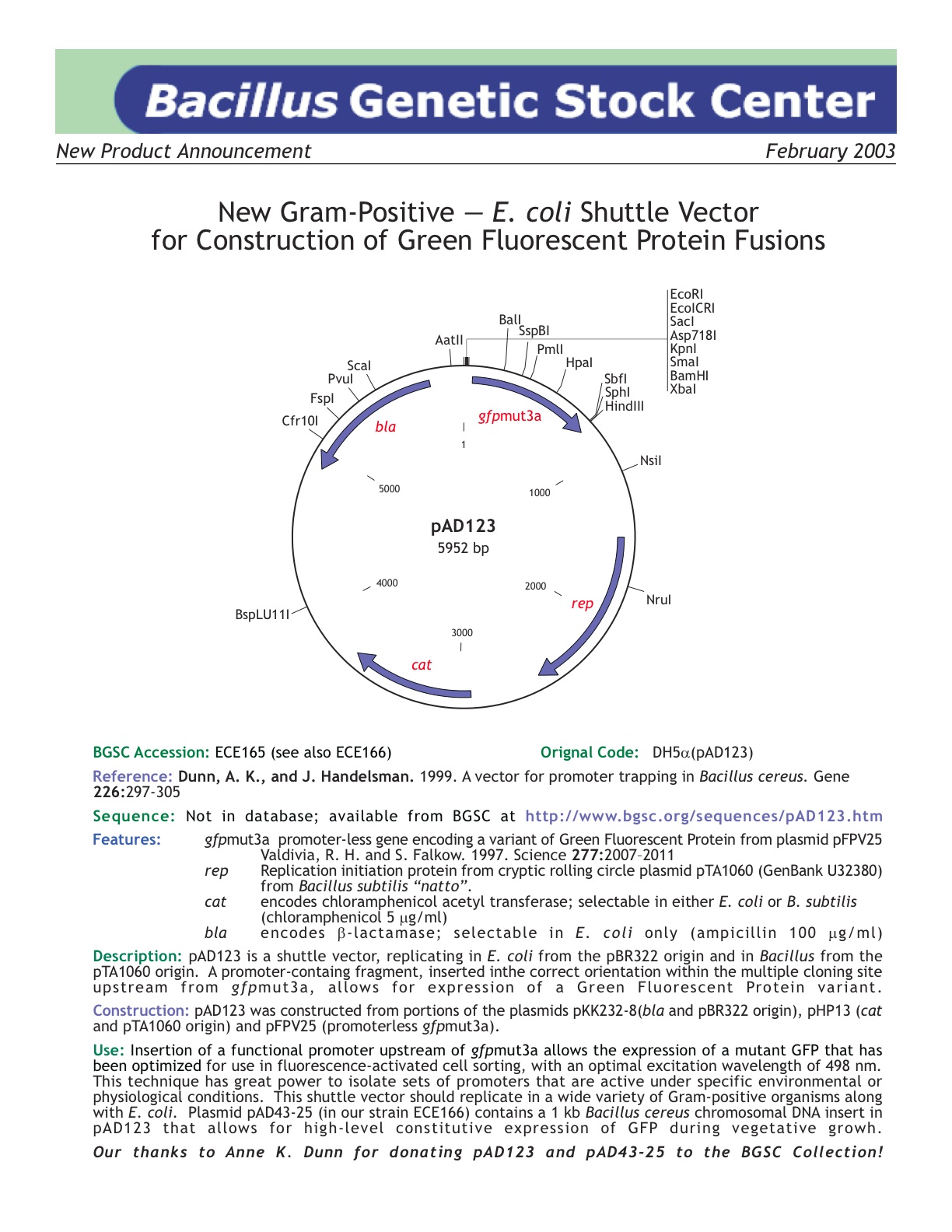
| ECE165 | pAD123 |
| [http://www.bgsc.org/sequences/pad123.htm BGSC sequence] | Yes | |
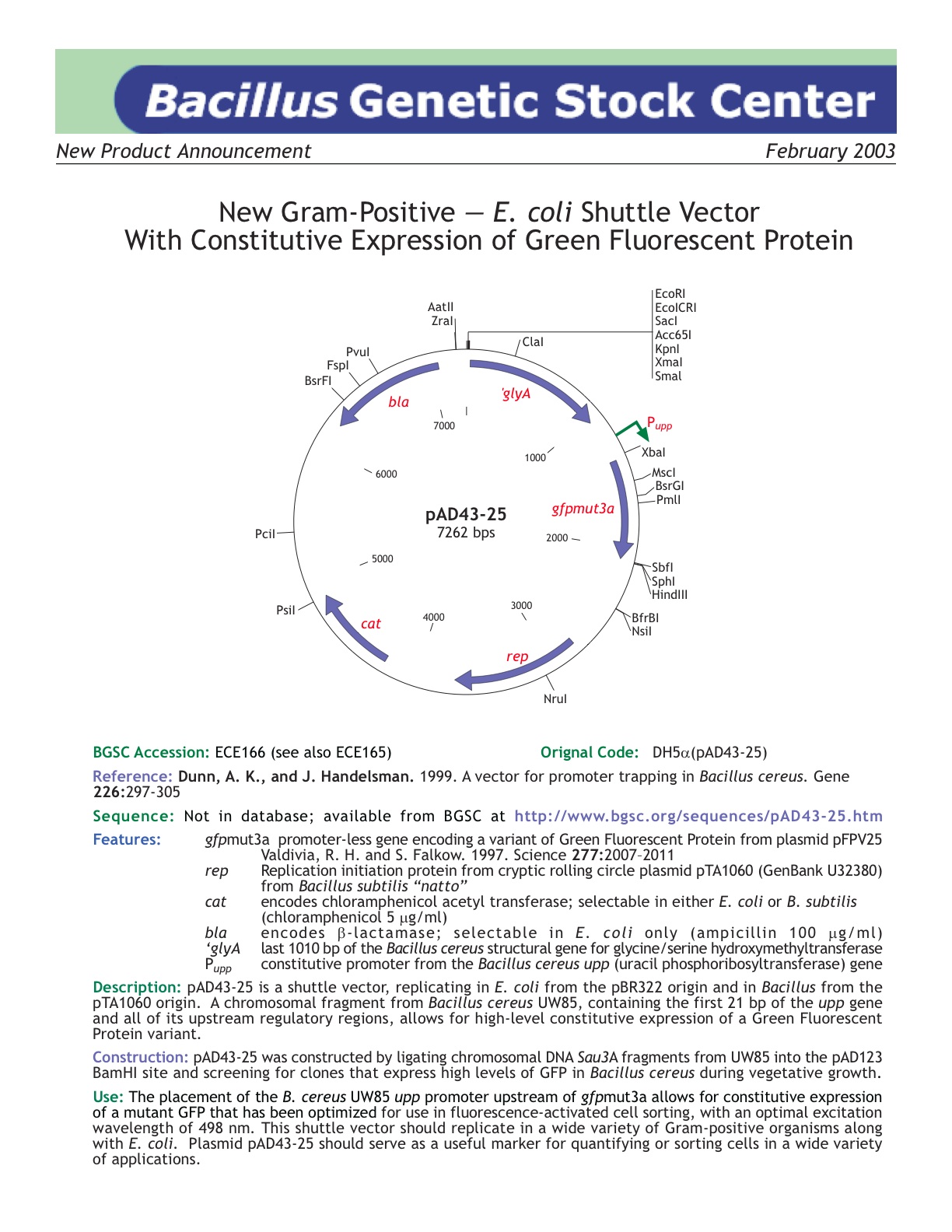
| ECE166 | pAD43-25 |
| [http://www.bgsc.org/sequences/pad43-25.htm BGSC sequence ] | Yes | |
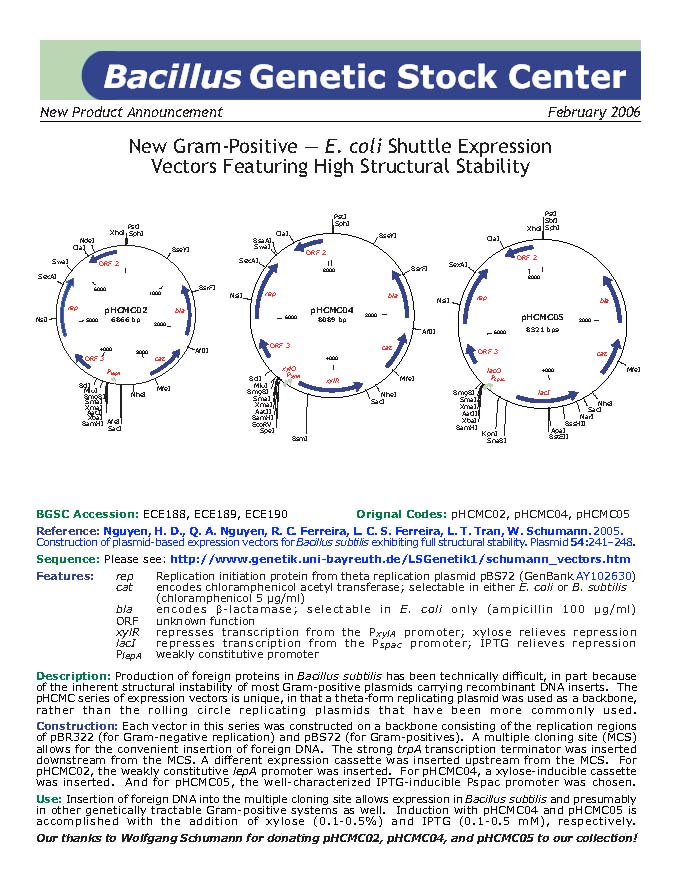
| ECE188 | pHCMC02 |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC02.htm From Bayreuth] | Not yet checked, DNA stocks | |
| ECE189 | pHCMC04 |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC04.htm From Bayreuth] | Not yet checked, DNA stocks | |
| ECE190 | pHCMC05 |
| [http://www.genetik.uni-bayreuth.de/LSGenetik1/schumann_pHCMC05.htm From Bayreuth] | Yes |
Plasmid Gels
- We ran these plasmids on gels, using double and single digests, to check them
 "
"