Team:Freiburg Transfection and Synthetic Receptor
From 2008.igem.org
(Difference between revisions)
| Line 13: | Line 13: | ||
<br> | <br> | ||
==Results== | ==Results== | ||
| - | < | + | <h4>'''Testing the transfection protocol'''</h4> |
| - | In order to check if the transfection protocol is suitable to transfect 293T cells a 'test'transfection with a lac z gene was done and the β-galactosidase was detected with an ONPG test.<br> | + | In order to check if the transfection protocol (Ca2+ precipitation) is suitable to transfect 293T cells a 'test'transfection with a lac z gene (Test-vector from Katja Arndt's lab) was done and the β-galactosidase was detected with an ONPG test.<br> |
<br> | <br> | ||
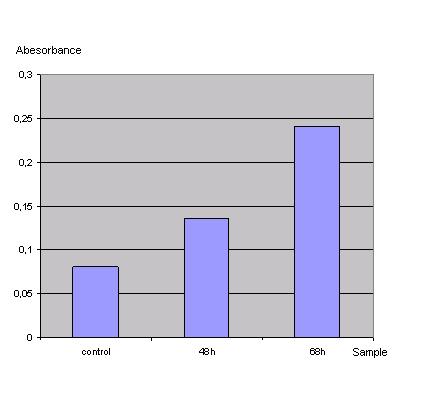
ONPG-assay: <br> | ONPG-assay: <br> | ||
| Line 22: | Line 22: | ||
''Graph 1_Transfection: Absorbance of o-Nitrophenol produced by the β-galactosidase (Control was done with untransfected cells using the same procedure)''<br> | ''Graph 1_Transfection: Absorbance of o-Nitrophenol produced by the β-galactosidase (Control was done with untransfected cells using the same procedure)''<br> | ||
<br> | <br> | ||
| - | < | + | Due to the results of the ONPG assay the Ca2+ precipitation proofed to be a very simple and effective method to transfect the 293t cells.<br> |
| - | To test the functionality of the transfectionvector and the CMV-promoter, YFP was cloned behind the promoter and the plasmid was brought into 293T cells. The detection of YFP took place 1 day later under a microscope with YFP filter.<br> | + | <br> |
| + | <h4>'''Testing the transfectionvector-CMV promoter construct'''</h4> | ||
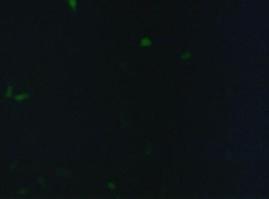
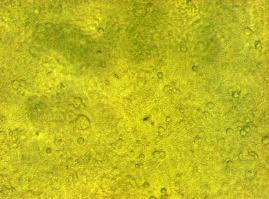
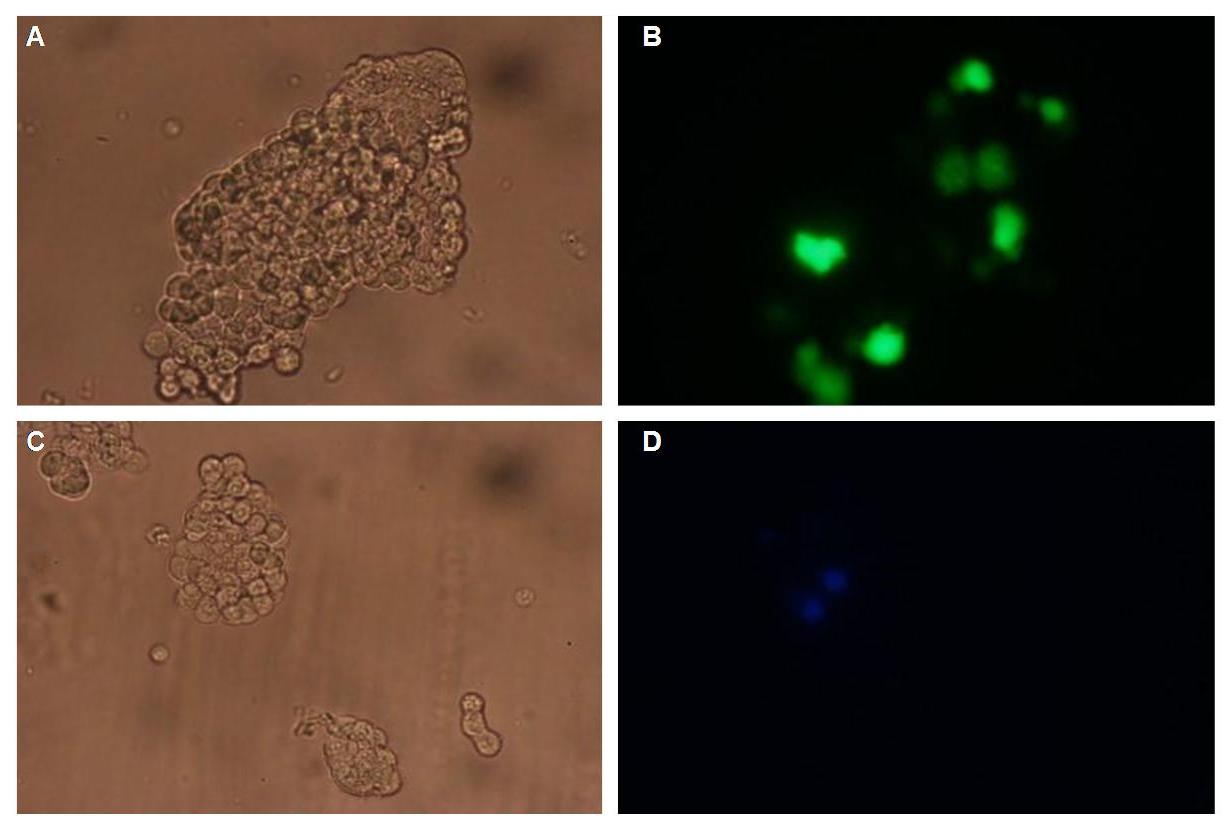
| + | To test the functionality of the transfectionvector and the CMV-promoter (BBa_K157040), YFP was cloned behind the promoter and the plasmid was brought into 293T cells. The detection of YFP took place 1 day later under a microscope with YFP filter.<br> | ||
The transfected cells show fluorescence by excitation of 510-520nm while the untransfected remain dark at this wavelength <br> | The transfected cells show fluorescence by excitation of 510-520nm while the untransfected remain dark at this wavelength <br> | ||
<br> | <br> | ||
| Line 29: | Line 31: | ||
''Figure 1_Transfection: transfected 293T cells (left), untransfected cells (middle) and transfected cells transmitted light (right)''<br> | ''Figure 1_Transfection: transfected 293T cells (left), untransfected cells (middle) and transfected cells transmitted light (right)''<br> | ||
<br> | <br> | ||
| - | < | + | <h4>'''Localization at the cell membrane'''</h4> |
To show the localization of the constructs at the cell membrane transfection of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP was performed.<br> | To show the localization of the constructs at the cell membrane transfection of the construct signalpeptide-Lipocalin-transmembraneregion-betaLactamase1-YFP was performed.<br> | ||
| - | Figure 2_Transfection shows the | + | Figure 2_Transfection shows the model of the protein structure based on PDB files. Anti-fluorescein Anticalin is the extracellular part of the construct. The transmembrane region is identical to that of the EGF-receptor erbb1 (BBa_K157002). Split-beta-Lactamase (BBa_, the intracellular part is labeled to the yellow fluorescent protein to detect membrane localization.<br> |
<br> | <br> | ||
[[Image:Freiburg2008_Lipo_bla1+YFP.jpg|500px]] | [[Image:Freiburg2008_Lipo_bla1+YFP.jpg|500px]] | ||
| Line 47: | Line 49: | ||
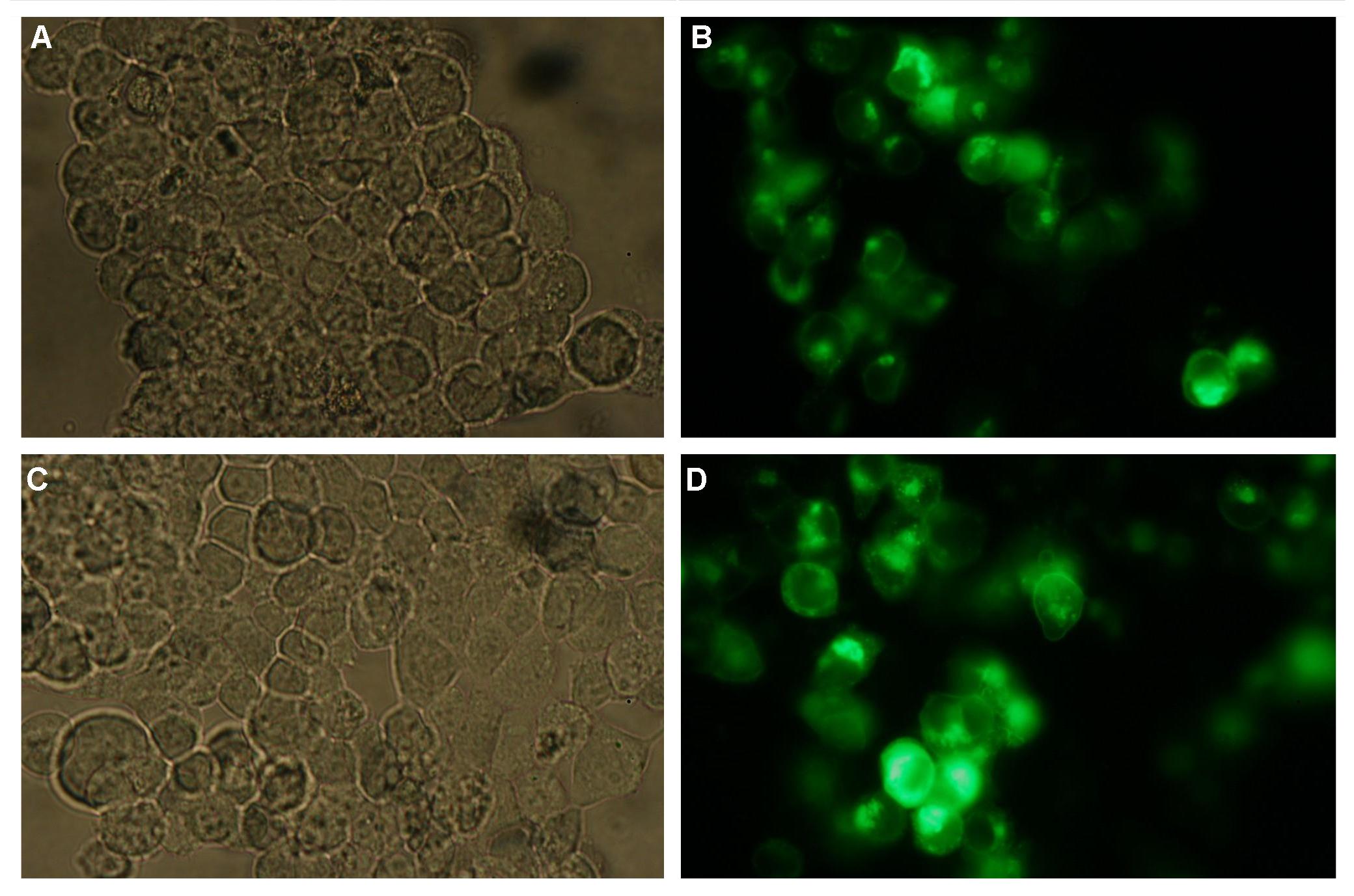
'''Figure 4_Transfection: 293T cells transfected with transfectionvector-CMV-YFP (A and B); 293T cells transfected with transfectionvector-CMV-CFP (C and D)'''<br> | '''Figure 4_Transfection: 293T cells transfected with transfectionvector-CMV-YFP (A and B); 293T cells transfected with transfectionvector-CMV-CFP (C and D)'''<br> | ||
<br> | <br> | ||
| - | < | + | <h4>'''Double transfections with Splitfluorophor-/Splitenzyme-constructs'''</h4> |
On Figure 5_Transfection the structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs are visible (exemplary for the Splitfluorophore-/Splitenzyme-constructs). The extracellular fragment is build of Lipocalin (fluorescein binding Anticalin) and a GGGSLinker. Intracellular either the N-terminal part or the C-terminal part of the splitfluorophore is fused to the transmembrane region of the EGF-receptor. To achieve more flexibility and to support the assembly of the two splitfluorophore parts a fluolinker is fused in between the transmembrane region and the C-terminal part of the splitfluorophores.<br> | On Figure 5_Transfection the structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs are visible (exemplary for the Splitfluorophore-/Splitenzyme-constructs). The extracellular fragment is build of Lipocalin (fluorescein binding Anticalin) and a GGGSLinker. Intracellular either the N-terminal part or the C-terminal part of the splitfluorophore is fused to the transmembrane region of the EGF-receptor. To achieve more flexibility and to support the assembly of the two splitfluorophore parts a fluolinker is fused in between the transmembrane region and the C-terminal part of the splitfluorophores.<br> | ||
<br> | <br> | ||
| Line 53: | Line 55: | ||
'''Figure 5_Transfection: structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs'''<br> | '''Figure 5_Transfection: structures of the signalpeptide-Lipocalin-transmembraneregion-nCFP and signalpeptide-Lipocalin-transmembraneregion-fluolinker-cCFP constructs'''<br> | ||
<br> | <br> | ||
| - | + | We hypothesize that adding fluorescein-coupled molecules leads to a clustering of the Lipocalin constructs due to the fluorescein-Lipocalin-binding. | |
The clustering of the constructs in turn results in an assembly of the splitfluorophores or splitenzymes and therefore creates a functional protein (Figure 6_Transfection).<br> | The clustering of the constructs in turn results in an assembly of the splitfluorophores or splitenzymes and therefore creates a functional protein (Figure 6_Transfection).<br> | ||
<br> | <br> | ||
Revision as of 15:14, 29 October 2008
 "
"