Team:Paris
From 2008.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
<br> | <br> | ||
<center><html><div style="color:#275D96; font-size:2em;">The BacteriO'Clock</div></html></center> | <center><html><div style="color:#275D96; font-size:2em;">The BacteriO'Clock</div></html></center> | ||
| + | <br> | ||
<br> | <br> | ||
| Line 12: | Line 13: | ||
</html> | </html> | ||
</center> | </center> | ||
| + | <br> | ||
| + | <br> | ||
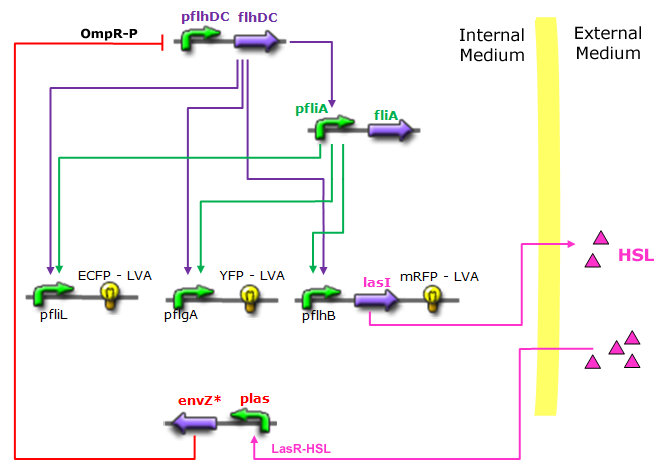
To achieve this incredible project, we relied on a well characterized genetic structure that allows a specific sequence of fluorescent proteins expression to occur in a First In - First Out order. This FIFO behaviour enables us to define a rich color encoding of day time. An additional negative feedback loop generates clock oscillations! | To achieve this incredible project, we relied on a well characterized genetic structure that allows a specific sequence of fluorescent proteins expression to occur in a First In - First Out order. This FIFO behaviour enables us to define a rich color encoding of day time. An additional negative feedback loop generates clock oscillations! | ||
| - | + | <br><br> | |
Based on in-depth studies and experimentally measured parameters we develope models that show that this system is not likely to oscillate, and indicate ways to improve the initial design. In particular, this can be achieved by HSL production that enforces a necessary delay and provides an elegant cell synchronization mechanism : | Based on in-depth studies and experimentally measured parameters we develope models that show that this system is not likely to oscillate, and indicate ways to improve the initial design. In particular, this can be achieved by HSL production that enforces a necessary delay and provides an elegant cell synchronization mechanism : | ||
<center> | <center> | ||
Revision as of 01:28, 30 October 2008
 "
"