Team:ESBS-Strasbourg/Labeling
From 2008.igem.org
(Difference between revisions)
| Line 51: | Line 51: | ||
*Missing EcoRI, HindIII, NotI, NdeI, XhoI, RsrII, BamHI, NcoI, BglI, SpeI, XbaI, and PstI. | *Missing EcoRI, HindIII, NotI, NdeI, XhoI, RsrII, BamHI, NcoI, BglI, SpeI, XbaI, and PstI. | ||
*Except for 5' and 3' ends no significant sequence identity runs with Cerulean CFP (BBa_E2020) or to GFP S65T as found in O'Shea deletion strain collection (originally from plasmid pFA6-GFP(S65T)-His3MX6). | *Except for 5' and 3' ends no significant sequence identity runs with Cerulean CFP (BBa_E2020) or to GFP S65T as found in O'Shea deletion strain collection (originally from plasmid pFA6-GFP(S65T)-His3MX6). | ||
| + | __NOTOC__ | ||
Revision as of 08:28, 16 July 2008
Fluorescence labeling
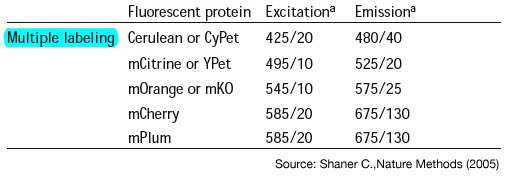
Multiple labeling
- [http://www.nature.com/nmeth/journal/v2/n12/abs/nmeth819.html;jsessionid=C9797F1441125016036E43C03175DD0D Shaner C.,Nature Methods (2005) -"A guide to choosing fluorescent proteins"]
- Propose cyan,yellow,orange and red => minimal crosstalk
- [http://www.clontech.com/products/detail.asp?product_id=10426&product_group_id=1437&product_family_id=1417&tabno=2 Reef Coral Fluorescent Proteins -Clontech]
- Said to be suitable for multiple labeling but forming tetramers (toxicity)
Features
| DNA binding domains | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | Excitation (nm) | Emission (nm) | taken from | reference | sequence available | work experimentally | remarks | treated by |
| GFPmut3b | 501 | 511 | biobrick registry | BBa E0040 | yes | yes | Paul | |
| EYFP | 515 | 528 | biobrick registry | BBa Jb3001 | yes | yes | Paul | |
| ECFP | 439 | 476 | biobrick registry | BBa E0022 | yes | ? | sequence without IRES | Paul |
| Highly RFP | 584 | 607 | biobrick registry | BBa E1010 | yes | ? | Paul | |
| BFP | 456/467/448 | PDB | yes | ? | Paul | |||
| OFP | PDB | AY678265 | yes | ? | Paul | |||
| BFP | 382 | 440 | Marielle | yes | ? | plasmid | Paul | |
| RFP | 561 | 583 | Marielle | yes | ? | plasmid | Paul |
- [http://www.clontech.com/images/brochures/FL852680_FloPro_DDD.pdf List from Clontech]
Biobricks
FPs that are available from the registry in standardized Biobrick-form
Yeast specific:
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63001 BBa_J63001], [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2030 BBa_E2030] -enhanced version of EYFP, yeast-optimized YFP
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2020 BBa_E2020] -enhanced version of ECFP, yeast-optimized (works?)
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2050 BBa_E2050] -derivative of mRFP1, yeast-optimized (works?) => mOrange
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2060 BBa_E2060] -derivative of mRFP1, yeast-optimized (works?) => mCherry
Notes:
- Contains N- and C-terminal linker sequences (with homology to biobrick parts BBa_E2060 (mCherry), BBa_E2050 (mOrange), and BBa_E2020 (Cerulean CFP) to facilitate color swapping in yeast.
- Adds N-"MATSG" and "GSGTA"-C. to published amino acid sequence.
- Inserts a "V" after normal start M.
- Double TAATAA stop codon.
- Missing EcoRI, HindIII, NotI, NdeI, XhoI, RsrII, BamHI, NcoI, BglI, SpeI, XbaI, and PstI.
- Except for 5' and 3' ends no significant sequence identity runs with Cerulean CFP (BBa_E2020) or to GFP S65T as found in O'Shea deletion strain collection (originally from plasmid pFA6-GFP(S65T)-His3MX6).
 "
"