Team:ESBS-Strasbourg/Labeling
From 2008.igem.org
Fluorescence labeling
If one wants to work with multiple fluorescent proteins one has a great variety of XFPs to chose from. This choice is even more difficult if more than one XFP is needed.
The main issue is the overlapping of the different extinction/emission curves. Therefore one has to pay attention to the particular maxima.
For example for our system already for the first bit it is crucial to work with suitable fluorescent markers.
Thus the purpose of this site is to give an overview for working with multiple fluorescence markers and to ease the decision which to take.
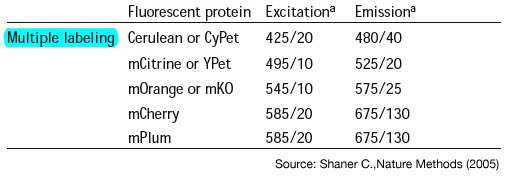
Multiple labeling
- [http://www.nature.com/nmeth/journal/v2/n12/abs/nmeth819.html;jsessionid=C9797F1441125016036E43C03175DD0D Shaner C.,Nature Methods (2005) -"A guide to choosing fluorescent proteins"]
- Propose cyan,yellow,orange and red => minimal crosstalk
- [http://www.clontech.com/products/detail.asp?product_id=10426&product_group_id=1437&product_family_id=1417&tabno=2 Reef Coral Fluorescent Proteins -Clontech]
- Said to be suitable for multiple labeling but forming tetramers (toxicity)
Features
| DNA binding domains | |||||||
|---|---|---|---|---|---|---|---|
| Name | Excitation (nm) | Emission (nm) | taken from | reference | sequence available | work experimentally | remarks |
| GFPmut3b | 501 | 511 | biobrick registry | BBa E0040 | yes | yes | |
| EYFP | 515 | 528 | biobrick registry | BBa Jb3001 | yes | yes | |
| ECFP | 439 | 476 | biobrick registry | BBa E0022 | yes | ? | sequence without IRES |
| Highly RFP | 584 | 607 | biobrick registry | BBa E1010 | yes | ? | |
| BFP | 456/467/448 | PDB | yes | ? | |||
| OFP | PDB | AY678265 | yes | ? | |||
| BFP | 382 | 440 | Lab | yes | ? | plasmid | |
| RFP | 561 | 583 | Lab | yes | ? | plasmid |
- [http://www.clontech.com/images/brochures/FL852680_FloPro_DDD.pdf List from Clontech] => Overview of available fluorescent proteins and their specifications
Biobricks
FPs that are available from the registry in standardized Biobrick-form
Yeast specific:
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63001 BBa_J63001], [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2030 BBa_E2030] -enhanced version of EYFP, yeast-optimized YFP
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2020 BBa_E2020] -enhanced version of ECFP, yeast-optimized (works?)
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2050 BBa_E2050] -derivative of mRFP1, yeast-optimized (works?) => mOrange
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_E2060 BBa_E2060] -derivative of mRFP1, yeast-optimized (works?) => mCherry
Notes:
- Contains N- and C-terminal linker sequences (with homology to biobrick parts BBa_E2060 (mCherry), BBa_E2050 (mOrange), and BBa_E2020 (Cerulean CFP) to facilitate color swapping in yeast.
- Adds N-"MATSG" and "GSGTA"-C. to published amino acid sequence.
- Inserts a "V" after normal start M.
- Double TAATAA stop codon.
- Missing EcoRI, HindIII, NotI, NdeI, XhoI, RsrII, BamHI, NcoI, BglI, SpeI, XbaI, and PstI.
- Except for 5' and 3' ends no significant sequence identity runs with Cerulean CFP (BBa_E2020) or to GFP S65T as found in O'Shea deletion strain collection (originally from plasmid pFA6-GFP(S65T)-His3MX6).
 "
"