Team:BCCS-Bristol/Modeling
From 2008.igem.org
(Difference between revisions)
Tgorochowski (Talk | contribs) (→Models) |
Tgorochowski (Talk | contribs) (→Models) |
||
| Line 32: | Line 32: | ||
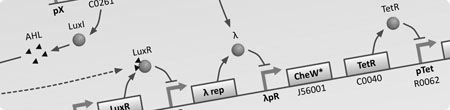
*[[Team:BCCS-Bristol/Modeling-GRN|'''Gene Regulatory Networks (GRNs)''']] - To understand the dynamics of genetic networks that we design differental equations are used to model the changes in concentrations of mRNA and proteins. | *[[Team:BCCS-Bristol/Modeling-GRN|'''Gene Regulatory Networks (GRNs)''']] - To understand the dynamics of genetic networks that we design differental equations are used to model the changes in concentrations of mRNA and proteins. | ||
| - | *[[Team:BCCS-Bristol/Modeling-Agent_Based|'''Stochastic Agent Based Simulation''']] | + | *[[Team:BCCS-Bristol/Modeling-Agent_Based|'''Stochastic Agent Based Simulation''']] - With the project requiring global behaviour to emerge via en masse movement of bacteria stochastic agent based simulations are used to investigate the viability and limits of possible methods. |
| - | *[[Team:BCCS-Bristol/Modeling-Hybrid|'''Hybrid Model''']] - With the aim of improving simulation accuracy this model | + | *[[Team:BCCS-Bristol/Modeling-Hybrid|'''Hybrid Model''']] - With the aim of improving simulation accuracy this model integrates the intercellular GRN dynamics with the extra-cellular interactions of the stochastic agent based simulation. |
==Results== | ==Results== | ||
Revision as of 20:39, 12 August 2008
|
Contents |
Approach
Modelling Notebook
- Progress Report - To track our progress see our weekly reports which let you know what we have been up to with comments on the reasons behind any important decisions.
- Modelling Parameters - Full listings of all parameters investigated for the modelling part of the project, with values that have been chosen, reasons for their selection and references to the original sources.
Models
- Gene Regulatory Networks (GRNs) - To understand the dynamics of genetic networks that we design differental equations are used to model the changes in concentrations of mRNA and proteins.
- Stochastic Agent Based Simulation - With the project requiring global behaviour to emerge via en masse movement of bacteria stochastic agent based simulations are used to investigate the viability and limits of possible methods.
- Hybrid Model - With the aim of improving simulation accuracy this model integrates the intercellular GRN dynamics with the extra-cellular interactions of the stochastic agent based simulation.
Results
Modelling Photo Album
|
|
 "
"