Team:Heidelberg/Notebook/Killing I/Notebook/week9
From 2008.igem.org
(Difference between revisions)
Maximilian.hoerner (Talk | contribs)
(New page: <html> <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/Heidelberg2.css' type='text/css' /> <link rel="stylesheet" href="http://igem-heidelberg.de/fileadmin/Wiki/Menu....)
Newer edit →
(New page: <html> <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/Heidelberg2.css' type='text/css' /> <link rel="stylesheet" href="http://igem-heidelberg.de/fileadmin/Wiki/Menu....)
Newer edit →
Revision as of 22:25, 25 October 2008


Week 9
Contents |
Monday, 09/29/08
new phage cloning strategy
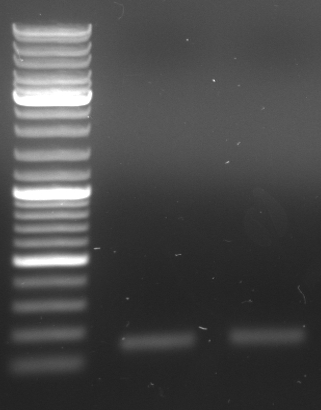
PCR of GFP (I20260) and terminator (B0015)
25µl Phusion Master Mix 2x 1µl GFP_new_fw 1µl GFP_new_rev 1µl Maxiprep I20260 (stock: 200ng/µl, dilution: 1:10) 22µl water ----- 50µl
25µl Phusion Master Mix 2x 1µl Term_new_fw 1µl Term_new_rev 1,5µl Miniprep B0015 (Andi, stock: 80ng/µl, dilution: 1:10) 21,5µl water ----- 50µl
PCR protocol 95°C 1min 95°C 30s | 55°C 30s | 26x 72°C 1min | 72°C 10min 4°C for ever
- each two pcr samples (GFP new 1, GFP new 2, Term new 1, Term new 2)
- Gel (1µl DNA)
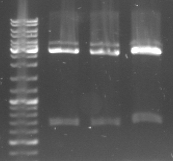
- expected sizes: GFP: 919bp + 28bp = 947bp, Terminator: 129bp + 22bp = 151bp
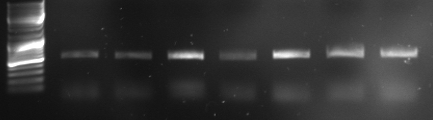
- lane 0: DNA ladder mix
- lane 1: Term new 1 (151bp)
- lane 2: Term new 2 (151bp)
- lane 0: DNA ladder mix
- lane 3: GFP new 1 (947bp)
- lane 4: GFP new 2 (947bp)
- -->PCR seems to be good
Processing of the KpnI Mutagenesis PCR from Friday
- DpnI digestion (1µl DpnI to 50µl PCR sample, 2h 37°C)
- PCR purification kit (eluted in 30µl water)
- Digestion with KpnI (2h 37°C)
2µl NEB1 2µl BSA 1µl DNA 2µl KpnI 13µl water ----- 20µl
- nothing can be seen on the gel --> do the digestion again with more template DNA
- Digestion with KpnI (1h 37°C)
5µl NEB1 5µl BSA 10µl DNA 3µl KpnI 27µl water ----- 50µl
- retrafo with 2µl and 6µl DNA on Amp/Cm plates
- no colonies could be seen on the next day
Cloning of oriT in standard plasmid
- inoculated colonies from transformation minipreped and digested with sfcI, one sample with NotI
- gel showed that we did not have the expected insert
- control PCR to verify this with ORiT primer and primer for standard plasmid -> no insert, too
- conclusion: start OriT cloning new
- OriT PCR with OriT primer with prefix and suffix (oriT_pre, oriT_suf) from pBluescript with insert using Phusion
- maxiprep of pSB1A3 with insert (T9002) inoculated
- Tomorrow: prep pSB1A3, digest with XbaI, PstI, cut backbone out of the gel, purificate, digest OriT with XbaI, PstI, ligate both and transform
- J01003 (oriT from registry) inoculated from glycerol stock to prep tomorrow and cotransform with pub307
Tuesday, 09/30/08
new cloning strategy
Mutagenesis PCR with mut_kpn1_pBlue
5 µl Pfu Buffer 5x 1 µl mut_kpn1_pBlue_fw 1 µl mut_kpn1_pBlue_rev 1,5 µl dNTPs 1 µl template DNA (pBlue with insert dilution: 1:200) 1 µl Pfu turbo 39,5 µl water ------ 50 µl
PCR protocol 95°C 30s 95°C 30s | 55°C 45s | 16x 68°C 14min | 4°C for ever
- 4 pcr samples
- 1µl DpnI to each sample--> 1h at 37°C
- PCR purification kit
- concentration measurement
- transformation in Top10 cells --> plated out on Cm plates
PCR of CmR
25µl Phusion Master Mix 2x 1µl CmR_new_fw 1µl CmR_new_rev 1µl Maxiprep pBlue with insert (stock: ~2µg/µl, dilution: 1:200) 22µl water ----- 50µl
PCR protocol 98°C 1min 98°C 10s | 61°C 10s | 26x 72°C 45s | 72°C 5min 4°C for ever
- 4 PCR samples
- PCR purification kit
- concentration measurement
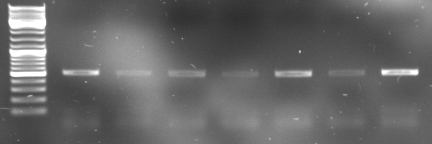
- Gel
- lane0: DNA ladder mix
- lane1-4: 1µl of pcr product (expected size: 738)
Cloning of CmR in pBluescript
- digestion of CmR pcr product (80ng/µl) with KpnI and HindIII
5µl NEB2 5µl BSA 10µl CmR 3µl KpnI 1,5µl HindIII 25,5µl water ----- 50µl
- digestion of the four pcr samples
- PCR purification kit
- digestion of Terminator pcr product (30ng/µl) with HindIII and SacI
5µl NEB2 5µl BSA 25µl Terminator 3µl SacI 1,5µl HindIII 10,5µl water ----- 50µl
- digestion of the two pcr samples
- PCR purification kit
- digestion of pBluescript (stock: maxiprep, 280ng/µl) with KpnI and SacI
5µl NEB1 5µl BSA 3µl pBluescript 2µl KpnI 2µl SacI 33µl water ----- 50µl
- digestion 4x
- Gel
- two bands: 2,9kb and 100bp
- cutted out 2,9kb band
- gel purification kit (2 samples: pBlue1 (106ng/µl) and pBlue2 (160ng/µl))
- Ligation (vector:insert = 1:3)
- each ligation one time with CmR1/Term1 and CmR2/Term2
- -->8 ligations
- ligations 30min at room temperature
- each ligation one time with CmR1/Term1 and CmR2/Term2
- Ligation (300ng, pBlue1):
pBlue: 1,4 µl CmR: 1,9 µl Term: 1 µl T4 Ligase: 1µl Buffer: 2µl water: 12,7µl
- Ligation (200ng, pBlue1):
pBlue: 1 µl CmR: 1,25 µl Term: 0,67 µl T4 Ligase: 1µl Buffer: 2µl water: 14,1µl
- Ligation (300ng, pBlue2):
pBlue: 1 µl CmR: 1,9 µl Term: 1 µl T4 Ligase: 1µl Buffer: 2µl water: 13,1µl
- Ligation (200ng, pBlue2):
pBlue: 0,6 µl CmR: 1,25 µl Term: 0,67 µl T4 Ligase: 1µl Buffer: 2µl water: 14,5µl
- transformation of the 8 ligations in TOP10, plated out on Cm plates
Cloning of oriT in standard plasmid
Producing the backbone
- maxiprep of T9002 in pSB1A3 (or pSB1A2 - will be sequenced by colicin group)
- Digestion with PstI, XbaI, NcoI for 1h (2 samples)
4µl NEB3 4µl BSA 2µl DNA 1µl PstI 1,5µl XbaI 1,5µl NcoI 26µl water ----- 40µl
- run gel and cut out 2000bp fragment, gel purification kit
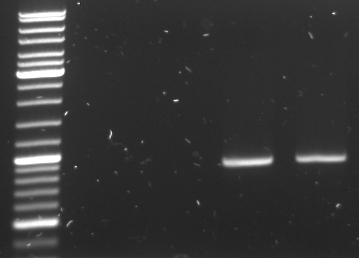
PCR of oriT
- first round of PCR amplification of OriT from pBluescript with insert went rong - no DNA in tubes
- second round worked, 3 samples made
25µl Phusion Master Mix 2x 2µl OriT_pref 2µl OriT_suf 1µl Maxiprep pBlue with insert (stock: ~2µg/µl, dilution: 1:200) 20µl water ----- 50µl
PCR protocol 98°C 1min 98°C 30s | 60°C 30s | 30x 72°C 45s | 72°C 5min 4°C for ever
Digestion and ligation
- digestion of oriT pcr product with PstI, XbaI
4µl NEB1 4µl BSA 28µl OriT 1,5µl PstI 1,5µl XbaI 1µl water ----- 40µl
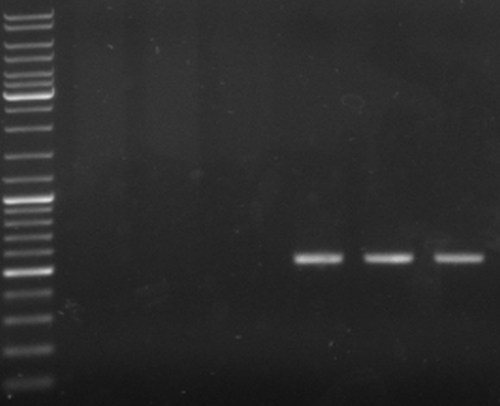
- check new oriT samples
- lane0: DNA ladder mix
- lane4-6: oriT pcr samples 1-3 (expected size: ~500bp)
- ligation: backbone to insert ratio 1:3, ligate for 40 min. at room temperature
4µl vector (backbone of T9002) 2µl insert (oriT PCR product) 1µl T4 Ligase 2µl Ligase Puffer 11µl water ----- 40µl
- transformation in TOP10
Conjugation Tests
Conjugation of pUB307 together with
- J01003 (2 samples)
- I714031 (2 samples)
- mix and let conjugate for 1h, plated in Kan/Amp plates
Test of J01003
- digestion with NcoI (to cut out OriT-insert) - worked
Wednesday, 10/01/08
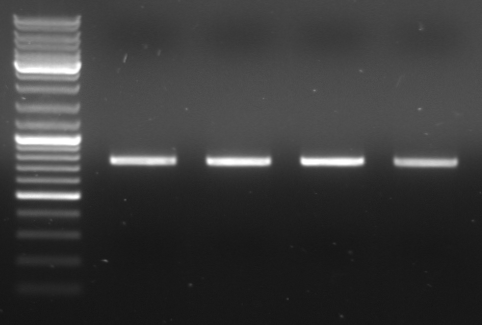
Cloning of oriT in standard plasmid
- transformation of oriT worked - colonies on all plates
- ligation tested with colony PCR - 2 colonies per plate
25µl PCR Master Mix 2x (Taq) 2µl VF2 2µl VR 21µl water
PCR protocol 95°C 5min 95°C 60s | 50.5°C 60s | 25x 72°C 60s | 4°C for ever
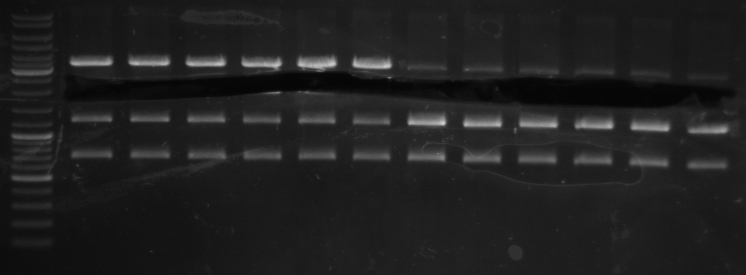
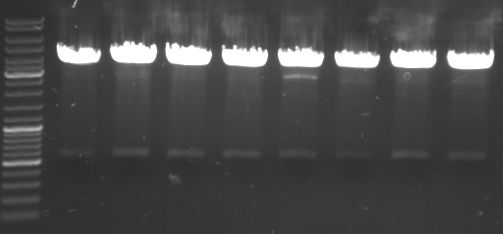
- gel results: all tested colonies had right insert (~600bp)
- top
- lane0: DNA ladder mix
- lane1-7: sample 1-7
- bottom
- lane0: DNA ladder mix
- lane1-6: sample 8-14
- sample 7 and 12 inoculated for maxiprep
oriT conjugation test
- samples 1, 7, 10, 12 conjugated with pUB307: take 2 ml of liquid culure, spin down and discard 1 ml supernatant, resuspend, mix cultures and plate on Kan/Amp
- conjugation of pUB307 in J01003 and I714031 worked
- liquid cultures for conjugation tests inoculated - did not grow
Lambda phage
- colony PCR of two samples (a and b) of last lambda ligation that gave colonies (and have already been minipreped:
- two PCR samples from each liquid culure
- gel result: expected fragment length: 4kb - all samples were positive!
cI
- cI received from geneart today
- digestion of cI (geneart) with EcoRI
- --> right fragment size
Proceeding of Mutagenesis PCR with mut_kpn1_pBlue
- inoculation of 12 overnight cultures of KpnI_mut_pBlue mutagenesis pcr colonies
Thursday, 10/02/08
Cloning of CmR in pBluescript
- on all 8 Cm plates of the pBlue+CmR+Terminator ligation colonies could be seen
- inoculation of 8 overnight cultures
Proceeding of Mutagenesis PCR with mut_kpn1_pBlue
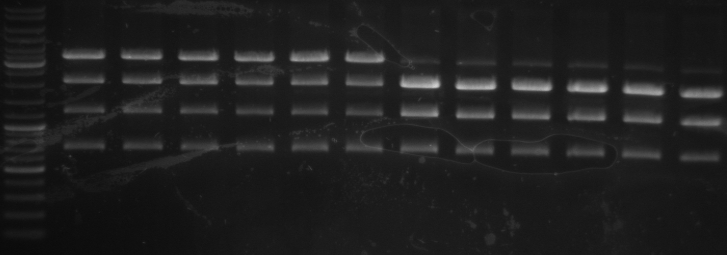
- 12 minipreps of KpnI mut1 pcr samples
Digestion with KpnI and AgeI to get the backbone
10µl DNA (~1µg) 2µl NEB1 2µl BSA 1µl KpnI 1µl AgeI 4µl water
- 12 samples
- Gel
- expected bands: 1.7kb and 5kb
- insert band with about 1.7kb on every sample
- 5kb and tight 6kb band
- cutted out both bands
Digestion of J01003 (oriT) to get the backbone pSB1A2
1µl DNA 5µl NEB3 5µl BSA 1,5µl PstI 2µl XbaI 35,5µl water
- 4 samples
- Gel
- 2kb and 1kb band
- -->cutted out 2kb band and purified
- Gel
pBlue Maxi 5.9 (doesn't make sense on gel) pBlue/insert 6kb pBlue/insert 6kb pBlue/insert 5kb pBlue/insert 5kb pSB1A2 2kb pSB1A2 2kb pcr product of insert from J01003 ladder
Friday, 10/03/08
Cloning of CmR in pBluescript
- miniprep of the 8 pBlue+CmR+Terminator overnight cultures
- concentrations [ng/µl]
- 189
- 216
- 261
- 273
- 264
- 192
- 270
- 239
- digestion of the Minipreps with KpnI and SacI
10µl Miniprep DNA 2µl NEB1 2µl BSA 1µl KpnI 1µl SacI 4µl water
- Gel
- lane0: DNA ladder mix
- lane1: 1
- lane2: 2
- lane3: 3a
- lane4: 3b
- lane5: 4
- lane6: 5
- lane7: 7
- lane8: 8
- gel overloaded
- one band at about 600bp, should be around 850
- proceeding: PCR with CmR primers, but they can't be found
- proceeding: sequencing of CmR in pBlue
- proceeding: digest again with a smaller amount of Miniprep DNA
- proceeding: digest with other enzymes to check if it's pBlue
Checking the insert of J01003
25µl Phusion Master Mix 1µl StandardFW 1µl StandardRV 1µl Maxiprep DNA (1:100) 22µl water
PCR protocol 95°C 30s 95°C 30s | 55°C 45s | 16x 68°C 14min | 4°C for ever
- insert has a length about 1,2kb
--> should be about 600bp
- maxiprep of two oriTs in pSB1A2 --> both were sequenced and digested with EcoRI/PstI
- result: oriT12 is T9002 (from sequencing) but oriT7 fits (digestion and sequencing) even though the sequencing with the forward primer was not succesful.
- our J01003 which we got from the MIT seems not to be correct!! After digestion with EcoRI/PstI there should appear a band at ~400bp but it runs at ~1200bp. This result was reproduced twice. Nevertheless it seems to be an oriT since it works succesfull in conjugation tests. We will prep it again next week from the glycerolstock and reepeat the digestion to make sure we did not contaminate it.
new phage cloning strategy
- digestion of GFP PCR product (70 ng/µl) with SacI and AgeI
10µl DNA 5µl NEB1 5µl BSA 2µl AgeI 2µl SacI 26µl water
- two digestion
- pcr purification kit
Sunday, 10/05/08
Cloning of CmR in pBluescript
- the cloning was done for a second time due to the bad digestion results on friday
- digestion of pBluescript with KpnI and SacI
5µl NEB1 5µl BSA 3µl pBluescript 1,5µl KpnI 1,5µl SacI 34µl water ----- 50µl
- two digestions
- Gel
- Gel extraction kit
- 8 overnight ligations of pBlue backbone, CmR3/CmR4 and Term1/2
- 16°C, 14h
 "
"