Team:Montreal/Notebook
From 2008.igem.org
(Difference between revisions)
(→Lab Protocols) |
(→Lab Protocols) |
||
| Line 13: | Line 13: | ||
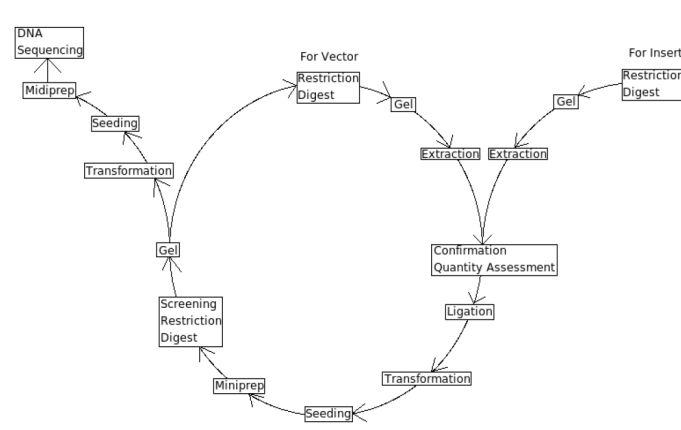
==Lab Protocols== | ==Lab Protocols== | ||
| + | Key Elements of the Central Dogma of cloning | ||
| + | 1. [https://2008.igem.org/Team:Montreal/Restriction_digest Restriction Digest] | ||
| - | + | 2. [https://2008.igem.org/Team:Montreal/DNA_Agarose_Gel Making a DNA Agarose Gel] | |
| - | + | 3. [https://2008.igem.org/Team:Montreal/Gel_extraction Gel extraction] | |
| - | + | 4. [https://2008.igem.org/Team:Montreal/Confirmation_quantity DNA quantity assessment] | |
| + | |||
| + | 5. [https://2008.igem.org/Team:Montreal/Ligation Ligation] | ||
| + | |||
| + | 6. [https://2008.igem.org/Team:Montreal/Cell_transformation Cell Transformation] | ||
| + | |||
| + | 7. [https://2008.igem.org/Team:Montreal/Seeding Seeding method] | ||
| + | |||
| + | 8. [https://2008.igem.org/Team:Montreal/Midiprep DNA Extraction: Mini-, Midi-, Maxiprep] | ||
==Lab Progress== | ==Lab Progress== | ||
Revision as of 03:19, 17 June 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
|
Lab Protocols
Key Elements of the Central Dogma of cloning 1. Restriction Digest
5. Ligation
8. DNA Extraction: Mini-, Midi-, Maxiprep
Lab Progress
May 21st, 2008:
- Restriction enzyme digest was done on the J-brick with EcoRI. Gel was run on the J-brick after the restriction digest. No DNA was detected on the gel (the ladder was visible on the gel).
May 22nd, 2008:
- Prepared TOP10 competent cells for eventual transformation.
- Performed Mini-prep on Reporter+ Cells
- Performed Digest and Gel on Reporter plasmid extract -- no DNA present, suggest follow-up maxi-prep
May 23rd, 2008:
- Transformed Top10 cells with Puc19 to ensure that the competent cell procedure was successful. Growth was observed, therefore procedure was successful.
May 25th, 2008:
- Diluted Reporter cells 1/1000 with 5ul Kan/mL culture for 16h incubation at 5:30pm. To be used for Maxiprep at 9:30am-1:30pm.
May 28th, 2008:
- Ran gel on Elowitz Reporter DNA cut with EcoR1; 2 bands
- 0.7 kb and 2.0 kb, confirms identity of reporter DNA.
- Seeded syn-I and J-40001 into amp/kan LB and kan LB.
May 29th, 2008:
- Growth of J-brick in culture - No growth of I-brick on culture
- Seeded J-brick for Midi-Prep in 40mL LB with ampicillin
- Transformed TOP10 cells with both I brick and Reporter Plasmid
June 2nd, 2008:
- Growth of I-brick on culture
- Midi-prep of both I and J brick followed by gel
- Gel indicates no presence of DNA, will be confirmed by spectrophotometric assay
June 3rd, 2008:
- Seeding of 5mL cultures of both I and J brick
- Identity of colonies on I brick plates is suspect, must ensure that eventual DNA gel confirms exact restriction digest
June 4th, 2008:
- Midiprep was done on the I and the J-brick. Once the isopropanol was added, the J-brick midiprep looked clear (no DNA was eluted). DNA gel needs to be done to confirm presence of DNA in both cases.
June 9th, 2008:
- Seeded J and I-brick re-seeded for Maxi-Prep
- Gel failed to confirm presence of previously collected DNA samples of J and I-brick, will be repeated following Maxi-Prep.
June 10th, 2008:
- Since no growth was observed in the I-brick culture, the I brick was re-seeded.
- The J-brick was diluted in 500-ml of LB broth(for a Maxiprep to be done the next day).
June 11th, 2008:
- Maxiprep of the J-brick.
- Restriction digest of J-brick.
- Dilution of I-brick in 500-ml of LB broth.
June 12th, 2008:
- Maxiprep of the I-brick.
- Restriction digest and gel of June 11th and June 12th I-brick and J-brick DNA using EcoR1. Bands revealed at roughly 4000bp and 2500bp for I-brick (expected 2652bp and 3939bp). J brick single band that was not informative, new digestion to be completed tomorrow.
June 16th, 2008:
- I-brick was seeded and diluted over last two days, but there was insufficient growth so it will be left to grow one more day before performing another midi-prep. This is to compliment the already successful Maxi-Prep that gave low concentrations of DNA.
- Another gel was performed of previous J-brick preps that confirmed the absence of the desired plasmid, no DNA was detected when digested with EcoR1.
- J-brick was re-transformed into TOP10 chemically competent cells and then plated on Amp+ plates.<ul/></p>
 "
"