Team:Alberta NINT/Modeling
From 2008.igem.org
(→Modeling) |
|||
| Line 19: | Line 19: | ||
=Modeling= | =Modeling= | ||
| - | + | Ability to model the folding of the designed RNA was desired not only to help visualize how the pieces would look, but also to investigate how interactions between the interactions between the inputs and outputs of the various genetic gates would play out. It is also important to examine possible interactions between the outputs of different gates as well as with other things that may be present in the cell. Reasonably accurate modeling could help lower cost of purchased DNA, save lab time, and result in a a more reliable final product. | |
| + | |||
| + | All modeling has been done using RNAStructure 4.5/4.6 (http://rna.urmc.rochester.edu/rnastructure.html) | ||
| + | |||
| + | Source code for this program was obtained with the hopes of making changes that would increase usability, as well as introduce new functions to the program that would allow it to be more useful for simulating some of the situations that may arise during the operation of the logic gate. | ||
| + | |||
| + | Currently, compiler version issues have prevented such changes from being implemented and tested. | ||
| + | |||
| + | <hr> | ||
| + | <br> | ||
| + | <br> | ||
| + | Windows access has been lacking for me for a few days now, so I played around with some single-stranded RNA folding. Definitely not the easiest thing to do... | ||
| + | |||
| + | The test sequence was folded in RNAStructure 4.6 run under Wine on a Linux system. The program used to generate the folds used a computational intelligence method to try and derive a probably fold. Refinements to the estimation of the folding energy of a structure have been made with the hopes of creating folds with some semblance of accuracy, but the problem may lie in the crossover/mutation operations being used in the evolutionary/genetic algorithm. | ||
| + | |||
| + | Pictures so I can feel as if I've accomplished something: | ||
| + | <center> | ||
| + | <gallery> | ||
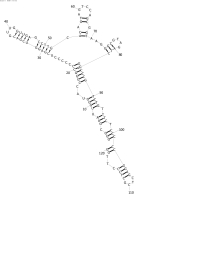
| + | Image:NINT_ControlFold.jpg|Control Fold Done with RNAStructure | ||
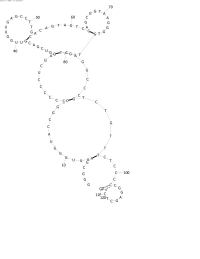
| + | Image:NINT SeriesA output1 scaled.jpg|Series A | ||
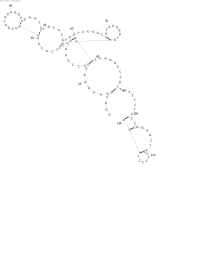
| + | Image:NINT_SeriesB_output1_scaled.jpg|Series B | ||
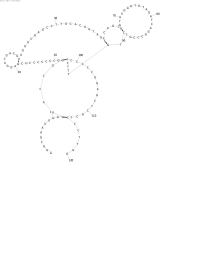
| + | Image:NINT_SeriesC_output1_scaled.jpg|Series C | ||
| + | Image:NINT_SeriesD_output1_scaled.jpg|Series D | ||
| + | |||
| + | </gallery> | ||
| + | </center> | ||
| + | <br> | ||
| + | The first few series were made trying improve the energy estimation subroutine. Series A was performed using only mutation and turbo simplistic energy rules. A bit of an refinement was made for Series B, as well as a change to the rate of mutation, which made it more dependent on the sequence length. Series C and D added a crossover operation to the mix, each was different in its treatment of bonds that had been broken at the point of crossing over. None of the series have exhibited any noticeable improvement over any of the others. | ||
| + | |||
| + | As viewed in the pictures, scaled for space, the common problem seems to be the creation of adjacent bonds (if that makes sense). Judging by the energy rules that are currently being employed, the problem likely resides in the EA. | ||
Revision as of 23:57, 22 June 2008
| Home | The Team | The Project | Lab Protocols | Bits and Pieces | Modeling | Notebook |
|---|
Modeling
Ability to model the folding of the designed RNA was desired not only to help visualize how the pieces would look, but also to investigate how interactions between the interactions between the inputs and outputs of the various genetic gates would play out. It is also important to examine possible interactions between the outputs of different gates as well as with other things that may be present in the cell. Reasonably accurate modeling could help lower cost of purchased DNA, save lab time, and result in a a more reliable final product.
All modeling has been done using RNAStructure 4.5/4.6 (http://rna.urmc.rochester.edu/rnastructure.html)
Source code for this program was obtained with the hopes of making changes that would increase usability, as well as introduce new functions to the program that would allow it to be more useful for simulating some of the situations that may arise during the operation of the logic gate.
Currently, compiler version issues have prevented such changes from being implemented and tested.
Windows access has been lacking for me for a few days now, so I played around with some single-stranded RNA folding. Definitely not the easiest thing to do...
The test sequence was folded in RNAStructure 4.6 run under Wine on a Linux system. The program used to generate the folds used a computational intelligence method to try and derive a probably fold. Refinements to the estimation of the folding energy of a structure have been made with the hopes of creating folds with some semblance of accuracy, but the problem may lie in the crossover/mutation operations being used in the evolutionary/genetic algorithm.
Pictures so I can feel as if I've accomplished something:
The first few series were made trying improve the energy estimation subroutine. Series A was performed using only mutation and turbo simplistic energy rules. A bit of an refinement was made for Series B, as well as a change to the rate of mutation, which made it more dependent on the sequence length. Series C and D added a crossover operation to the mix, each was different in its treatment of bonds that had been broken at the point of crossing over. None of the series have exhibited any noticeable improvement over any of the others.
As viewed in the pictures, scaled for space, the common problem seems to be the creation of adjacent bonds (if that makes sense). Judging by the energy rules that are currently being employed, the problem likely resides in the EA.
 "
"