Team:Heidelberg/Project/Visualization
From 2008.igem.org
(→Microscopic Screening and Quantification of 2D bacterial landscapes) |
(→Microscopic Screening and Quantification of 2D bacterial landscapes) |
||
| Line 481: | Line 481: | ||
Screening of bacterial density in soft agar chamber (visit [https://2008.igem.org/Team:Heidelberg/Notebook/visualization Notebook - Visualization] for more technical information). For a proof of principle the chemotactic ''E. coli'' strain HCB33 is applied to the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. | Screening of bacterial density in soft agar chamber (visit [https://2008.igem.org/Team:Heidelberg/Notebook/visualization Notebook - Visualization] for more technical information). For a proof of principle the chemotactic ''E. coli'' strain HCB33 is applied to the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. | ||
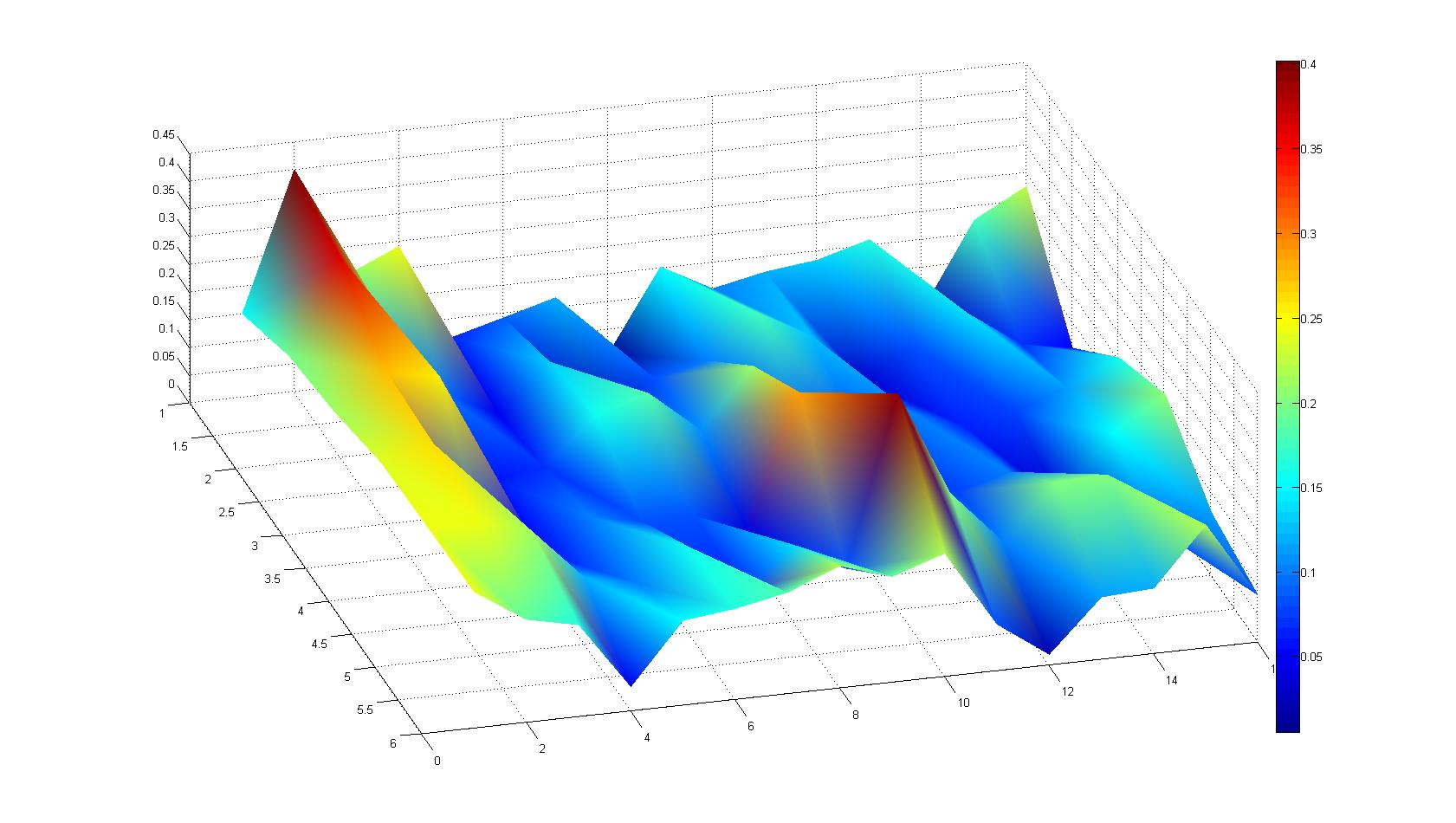
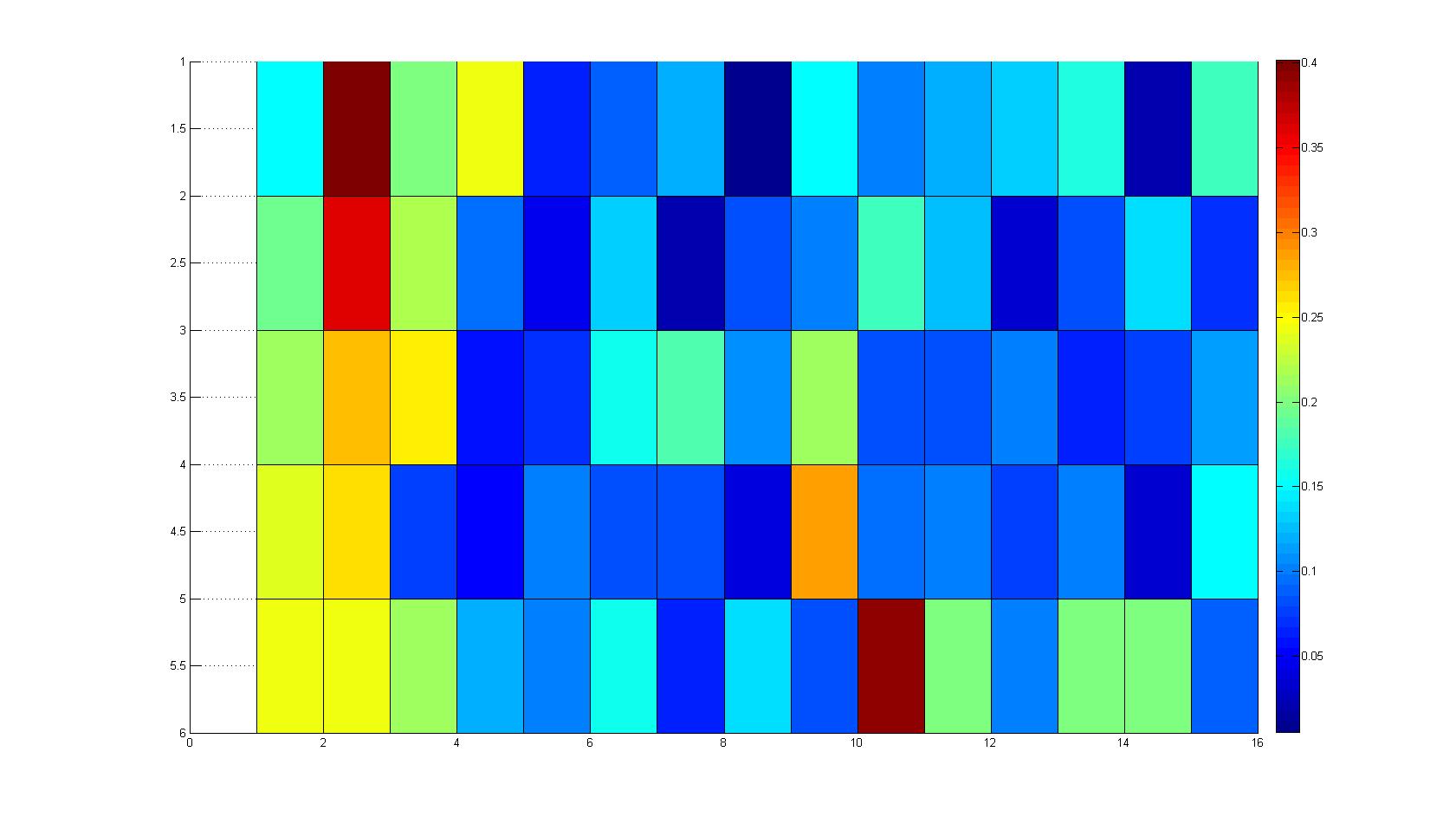
| - | After ... hours the bacteria accumulate around the plug, because they sense the established gradient of amino acids. Calculation of the mean of each image and subtraction of the background leads to a quantitative 2D representation of the bacterial density landscape. This data can be compared to 2D chemotactic modeling results based on a system of partial differential equations (visit [https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis Modeling - Chemotaxis/Colicins)] | + | After ... hours the bacteria accumulate around the plug, because they sense the established gradient of amino acids. Calculation of the mean of each image and subtraction of the background leads to a quantitative 2D representation of the bacterial density landscape. This data can be compared to 2D chemotactic modeling results based on a system of partial differential equations (visit [https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis Modeling - Chemotaxis/Colicins)]. Furthermore the data suggests ... |
Revision as of 18:17, 28 October 2008


Contents |
Killing - Colicin
Time series of killer and prey bacteria over 48 hours. Killer cells are marked with mCherry (red) and prey bacteria are marked with GFP (green). The intial ratio is 1:1, which can be recognized on the second and third frame (8h and 12h, respectively). Many prey and killer bacteria are grown after 36 hours (last three image frames). However, the faint green fluorescence (it is sharp!) indicates the weak prey metabolism due to the toxin. Toxin production and excretion is activated in killer bacteria by the signaling molecule autoinducer-1.
To see separate images click here
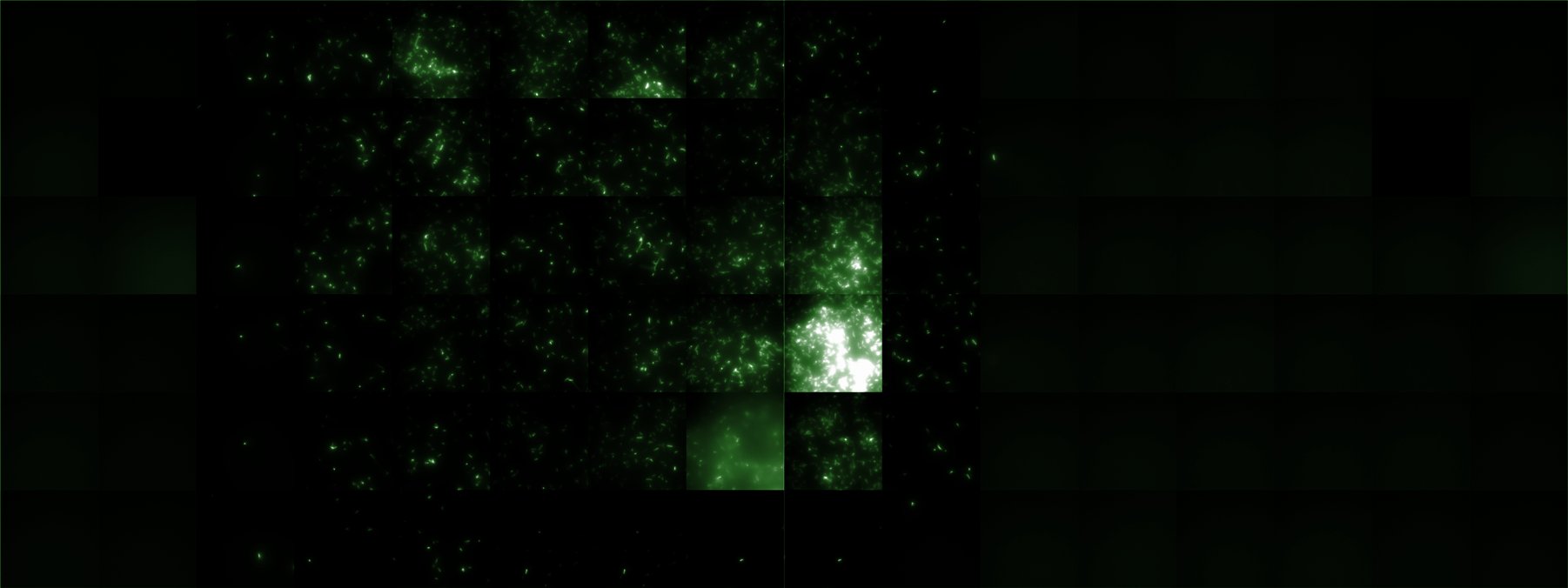
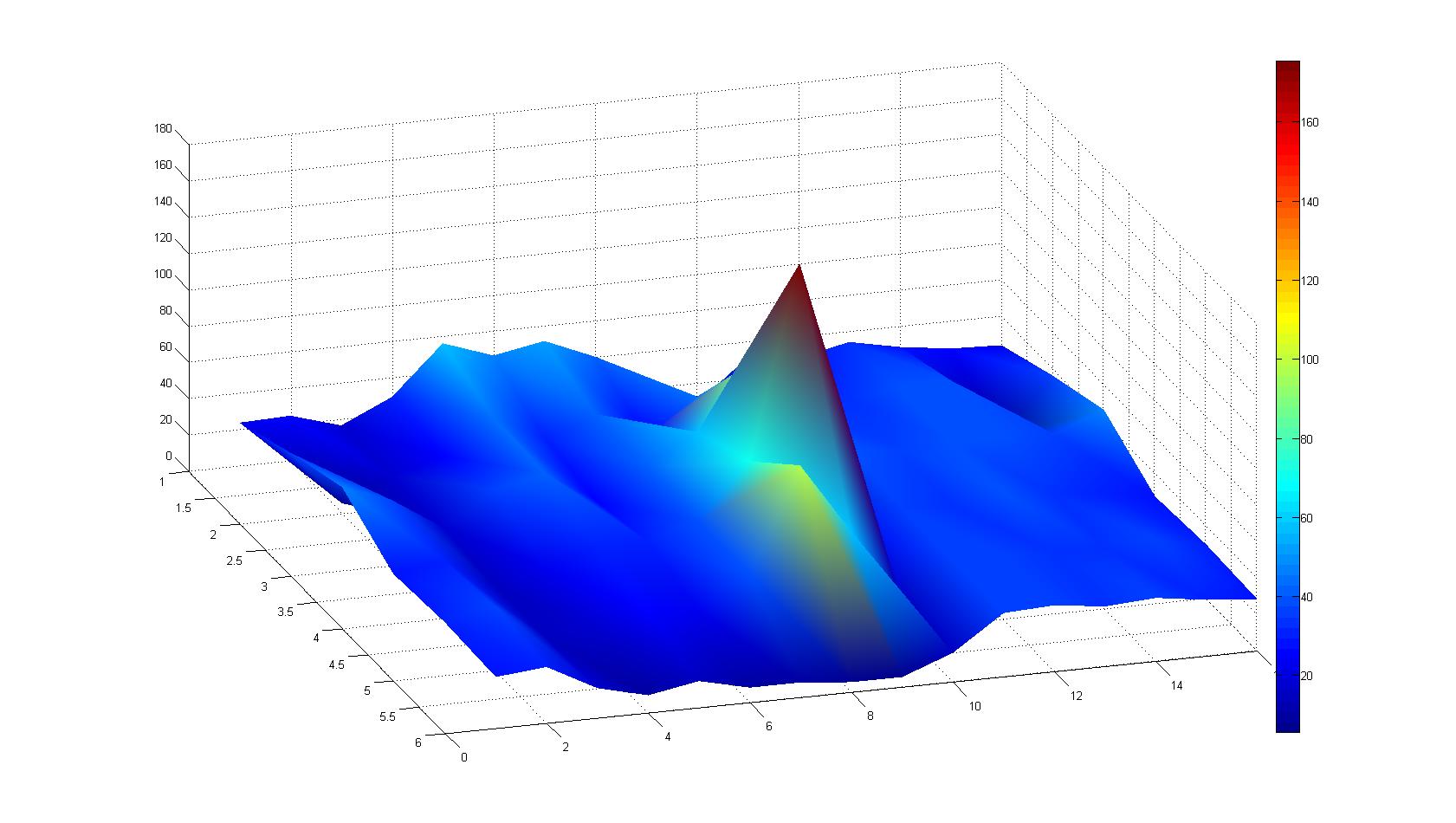
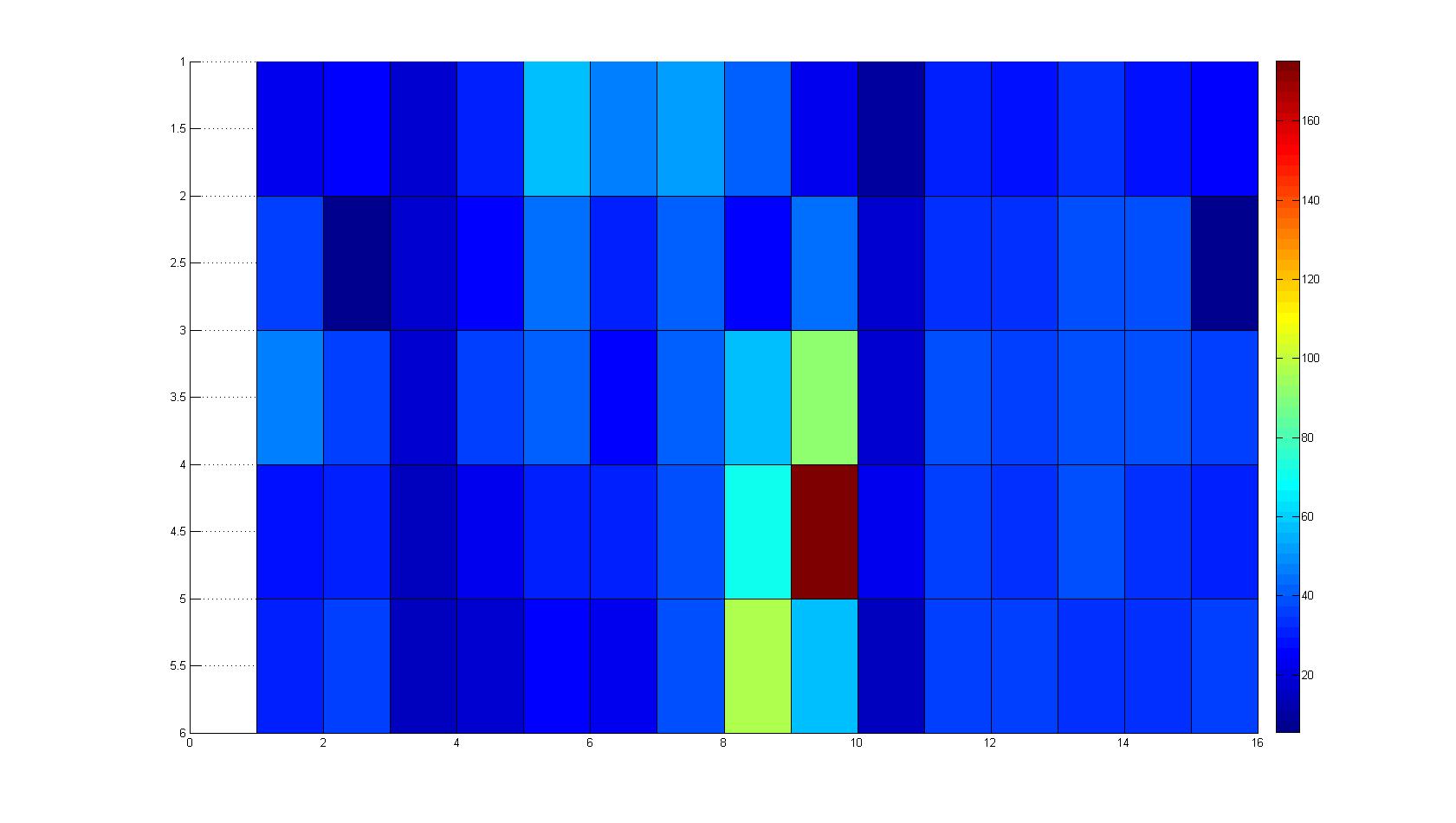
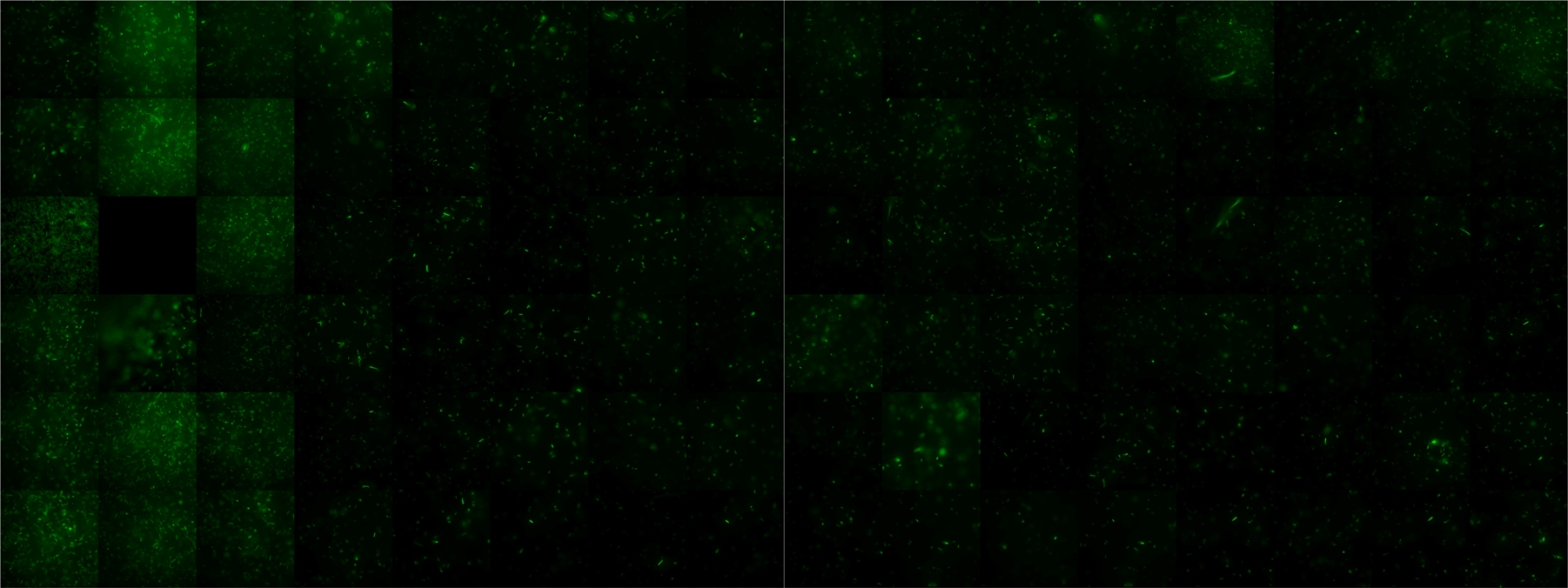
Microscopic Screening and Quantification of 2D bacterial landscapes
Screening of bacterial density in soft agar chamber (visit Notebook - Visualization for more technical information). For a proof of principle the chemotactic E. coli strain HCB33 is applied to the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. After ... hours the bacteria accumulate around the plug, because they sense the established gradient of amino acids. Calculation of the mean of each image and subtraction of the background leads to a quantitative 2D representation of the bacterial density landscape. This data can be compared to 2D chemotactic modeling results based on a system of partial differential equations (visit Modeling - Chemotaxis/Colicins). Furthermore the data suggests ...
Remark: The scans are taken from two different experiments with equal settings due to the difficult adjustment of a constant focus.
| After 0 hours | |
| After ? hours | |
 "
"