Team:Harvard/Shewie
From 2008.igem.org
(→Molecular Biology with Shewanella oneidensis) |
(→Molecular Biology with Shewanella oneidensis) |
||
| Line 67: | Line 67: | ||
{|align="justify" style="background-color:#FFFFFF;text-indent: 15pt; text-align:justify" width="100%" | {|align="justify" style="background-color:#FFFFFF;text-indent: 15pt; text-align:justify" width="100%" | ||
| | | | ||
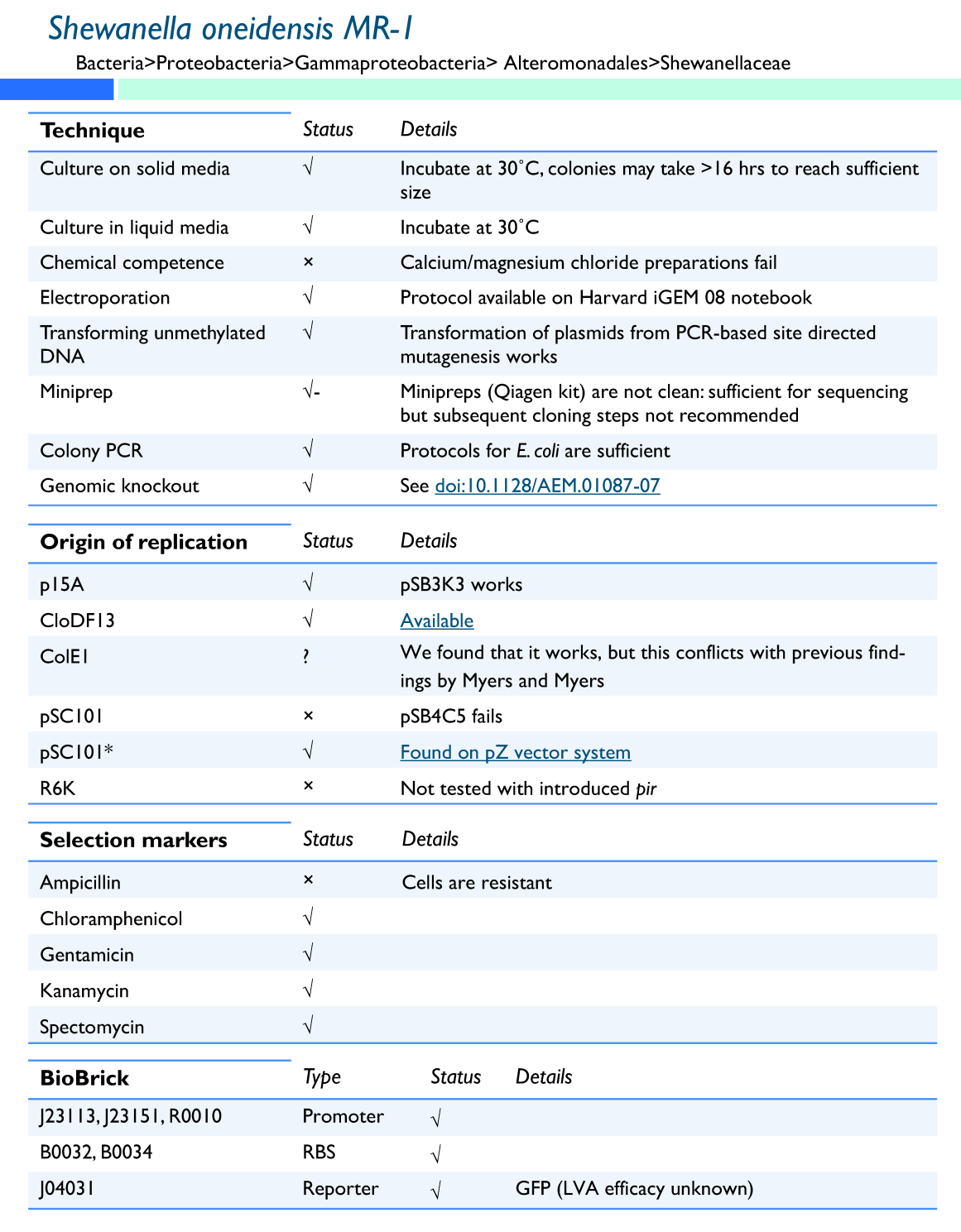
| - | The ''Shewanella oneidensis MR-1'' genome was sequenced in 2002, greatly increasing its usefulness as a model organism. It was found that it had a 4,969,803 base pair circular chromosome and a 161,613 base pair plasmid (Heidelberg et al. 2002). When cloning in ''S. oneidensis MR-1'', it has also been shown that plasmids with p15A origins replicate freely, whereas plasmids with a pMB1 origin of replication do not (Myers and Myers 1997). We further found that the pSC101* origin from Lutz and Bujard (2007) and the CloDF3 origin on the [http://www.emdbiosciences.com/html/NVG/DuetTable.html| pCDF-Duet vector] from Novagen work in ''Shewanella''. ''S. oneidensis MR-1'' grows at 30 ºC, can be electroporated (see protocol in our [[Team:Harvard/GenProtocols| Notebook]]) and forms round orange pink colonies on plates. It is resistant to ampicillin, but other resistance markers | + | The ''Shewanella oneidensis MR-1'' genome was sequenced in 2002, greatly increasing its usefulness as a model organism. It was found that it had a 4,969,803 base pair circular chromosome and a 161,613 base pair plasmid (Heidelberg et al. 2002). When cloning in ''S. oneidensis MR-1'', it has also been shown that plasmids with p15A origins replicate freely, whereas plasmids with a pMB1 origin of replication do not (Myers and Myers 1997). We further found that the pSC101* origin from Lutz and Bujard (2007) and the CloDF3 origin on the [http://www.emdbiosciences.com/html/NVG/DuetTable.html| pCDF-Duet vector] from Novagen work in ''Shewanella''. However, they are not pir+, so the R6K pir+ dependent origin does not work for them. ''S. oneidensis MR-1'' grows at 30 ºC, can be electroporated (see protocol in our [[Team:Harvard/GenProtocols| Notebook]]) and forms round orange pink colonies on plates. It is resistant to ampicillin, but other resistance markers, such as gentamycin and spectomycin, can be used (Saffarini). Together, these characteristics make ''S. oneidensis MR-1'' a genetically tractable organism good for exploring the possibilities of regulated bacterial electrical output. |
|<div style="text-indent:0pt;color:black">[[Image:Shewanella colonies growing on plate.JPG|thumb|300px|''S. oneidensis MR-1'' colonies from a transformation]]</div> | |<div style="text-indent:0pt;color:black">[[Image:Shewanella colonies growing on plate.JPG|thumb|300px|''S. oneidensis MR-1'' colonies from a transformation]]</div> | ||
|- | |- | ||
Revision as of 05:11, 29 October 2008
|
|
 "
"