Team:Rice University/Notebook/7 June 2008
From 2008.igem.org
(Difference between revisions)
StevensonT (Talk | contribs) |
StevensonT (Talk | contribs) |
||
| Line 4: | Line 4: | ||
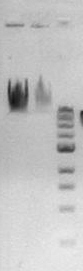
**Prepared gel electrophoresis of above digest along with solution of lambda DNA with 1Kb standard | **Prepared gel electrophoresis of above digest along with solution of lambda DNA with 1Kb standard | ||
[[Image:20080607_gel.jpg|100px|left]] | [[Image:20080607_gel.jpg|100px|left]] | ||
| - | (from | + | (from right to left, lane 1: 1Kb, lane 2: lambda DNA, lane 3: lambda with XhoI) |
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
*Taylor Stevenson | *Taylor Stevenson | ||
Latest revision as of 19:15, 25 June 2008
Saturday 7 June
- Selim Sheikh:
- Prepared digestion of lambda DNA with XhoI only
- Prepared gel electrophoresis of above digest along with solution of lambda DNA with 1Kb standard
(from right to left, lane 1: 1Kb, lane 2: lambda DNA, lane 3: lambda with XhoI)
- Taylor Stevenson
- Phage infected XL1-blue MR colonies cultured in 96 deep-well plate were spotted onto two EMB plates using a 2uL plate replicator. Both plates were incubated O/N, one at 30*C and one at 42*C (temp is high enough to denature the CI repressor, causing any lysogen to become lytic).
- Result-both plates showed no bacterial growth after 24h.
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"