Team:Heidelberg/Project/Visualization
From 2008.igem.org
(→After 0 hours) |
(→Microscopic Scanning and Quantification of 2D bacterial landscapes) |
||
| Line 482: | Line 482: | ||
A scan of the bacterial density in a soft agar chamber (visit [https://2008.igem.org/Team:Heidelberg/Notebook/visualization Notebook - Visualization] for more technical information). For a proof of principle the chemotactic ''E. coli'' strain HCB33 is applied in the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. | A scan of the bacterial density in a soft agar chamber (visit [https://2008.igem.org/Team:Heidelberg/Notebook/visualization Notebook - Visualization] for more technical information). For a proof of principle the chemotactic ''E. coli'' strain HCB33 is applied in the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. | ||
| - | After | + | After 21 hours the bacteria accumulate around the plug, because they sense the established gradient of amino acids. Calculation of the mean of each image and subtraction of the background leads to a quantitative 2D representation of the bacterial density landscape. This data can be compared to the 2D chemotactic modeling results based on a system of partial differential equations (visit [https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis Modeling - Chemotaxis/Colicins)]. |
Remark: The scans are taken from two different experiments with equal settings due to the difficult adjustment of a constant focus. | Remark: The scans are taken from two different experiments with equal settings due to the difficult adjustment of a constant focus. | ||
Revision as of 12:09, 29 October 2008


Contents |
Killing - Colicin
Time series of killer and prey bacteria over 48 hours. Killer cells are marked with mCherry (red) and prey bacteria are marked with GFP (green). The intial ratio is 1:1, which can be recognized on the second and third frame (8h and 12h, respectively). Many prey and killer bacteria are grown after 38 hours (last three image frames). However, the faint green fluorescence (it is sharp!) indicates the weak prey metabolism due to the toxin. Toxin production and excretion is activated in killer bacteria by the signaling molecule autoinducer-1 (visit Project - Colicins for more biological information).
For viewing this video, the [http://www.apple.com/quicktime/download/| QickTime Player] (free on apple.com) is required. To see separate images click here
[back]
Microscopic Scanning and Quantification of 2D bacterial landscapes
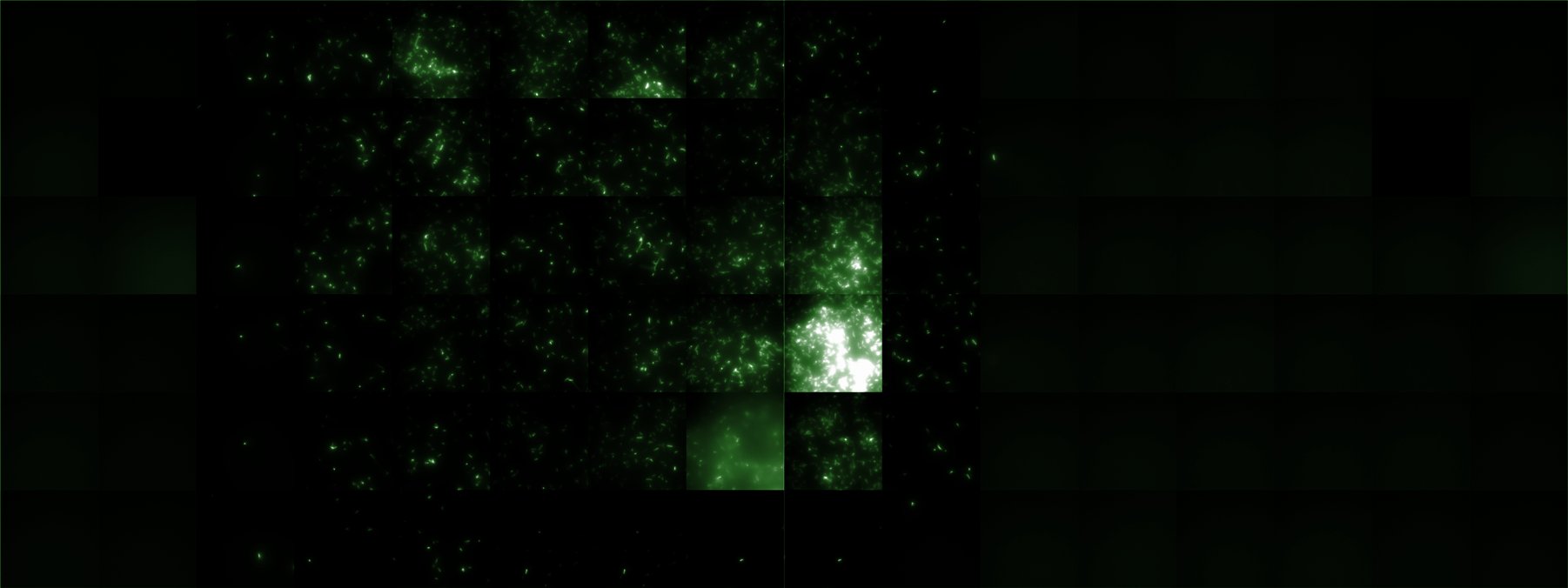
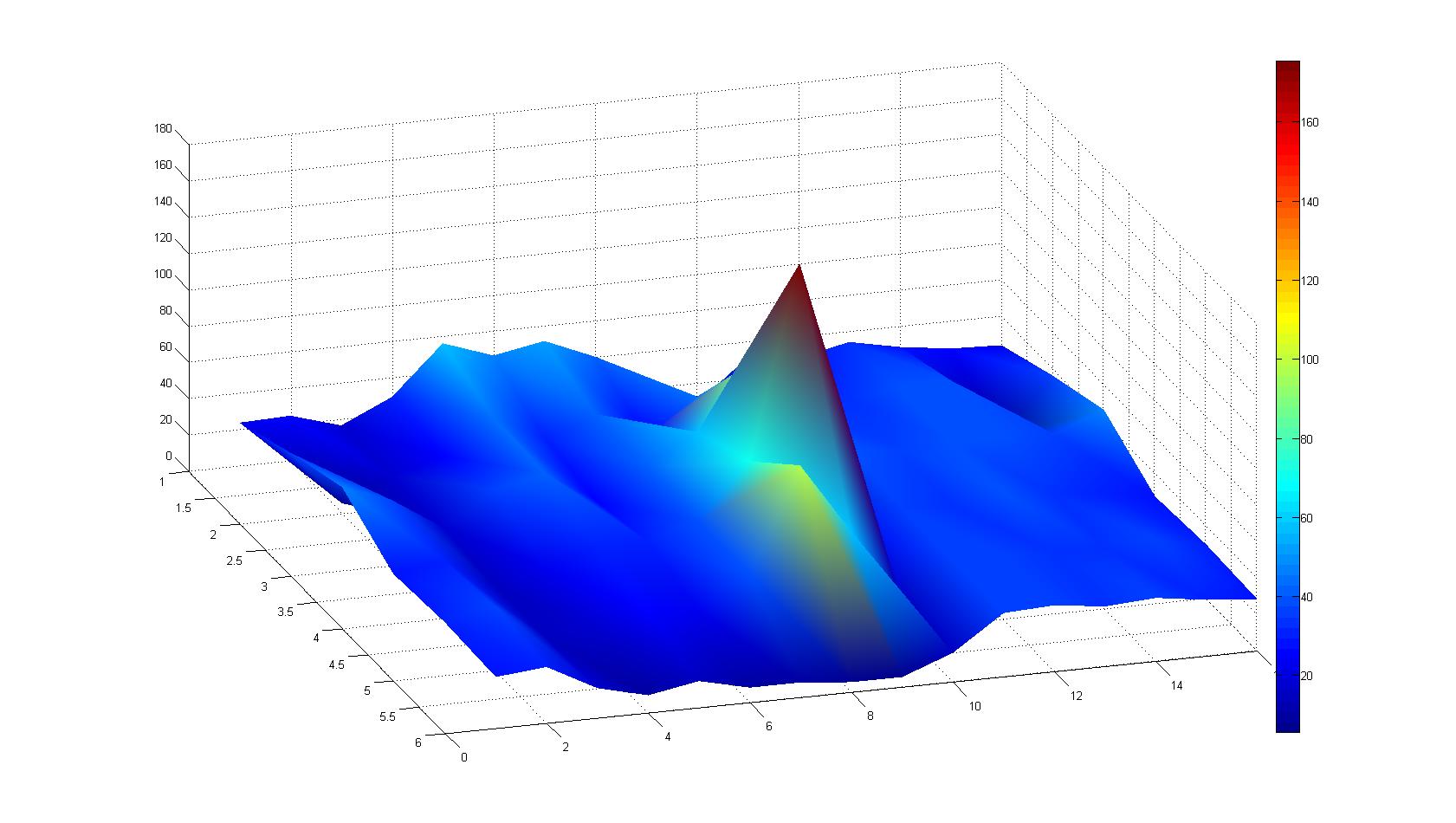
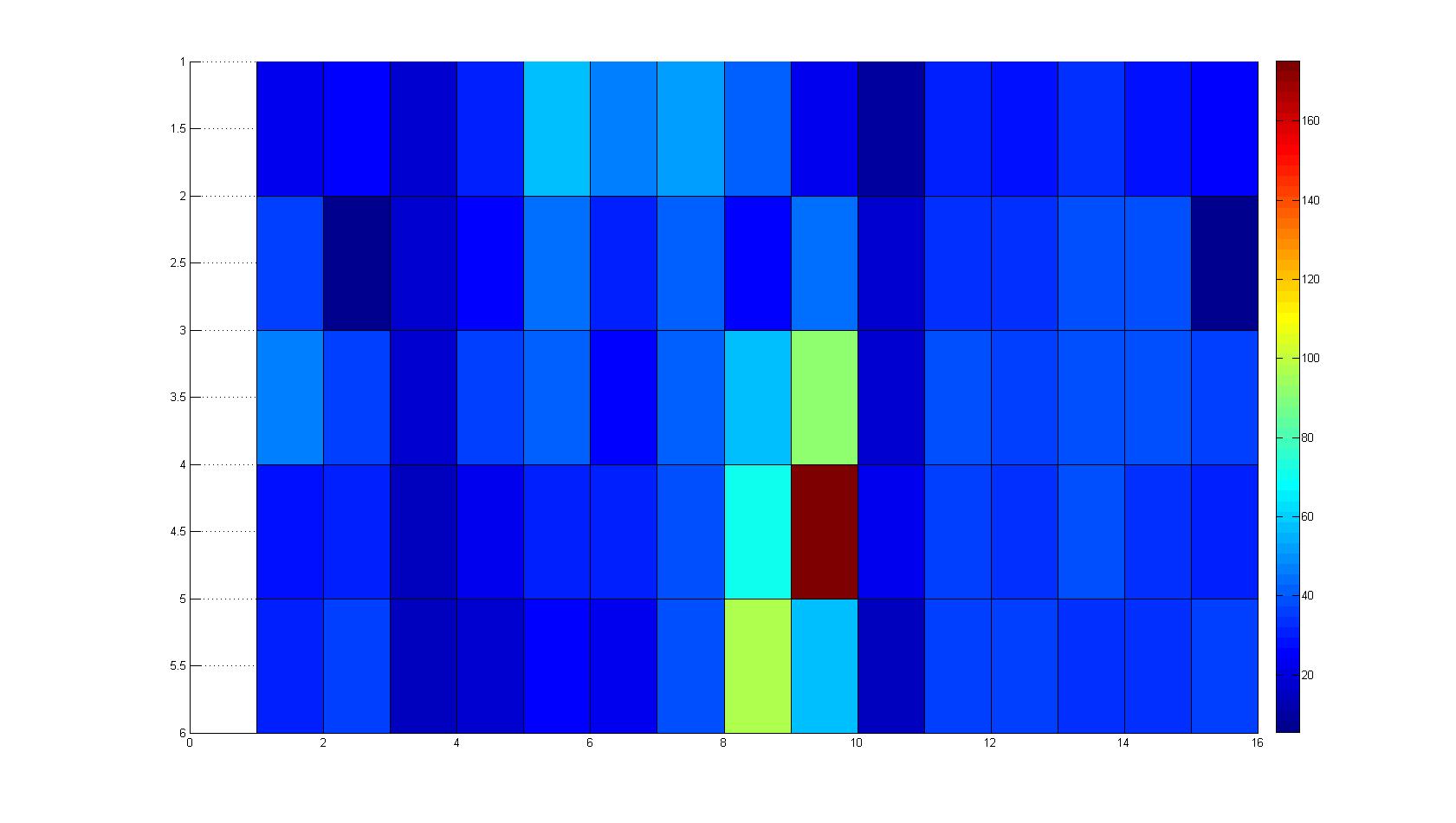
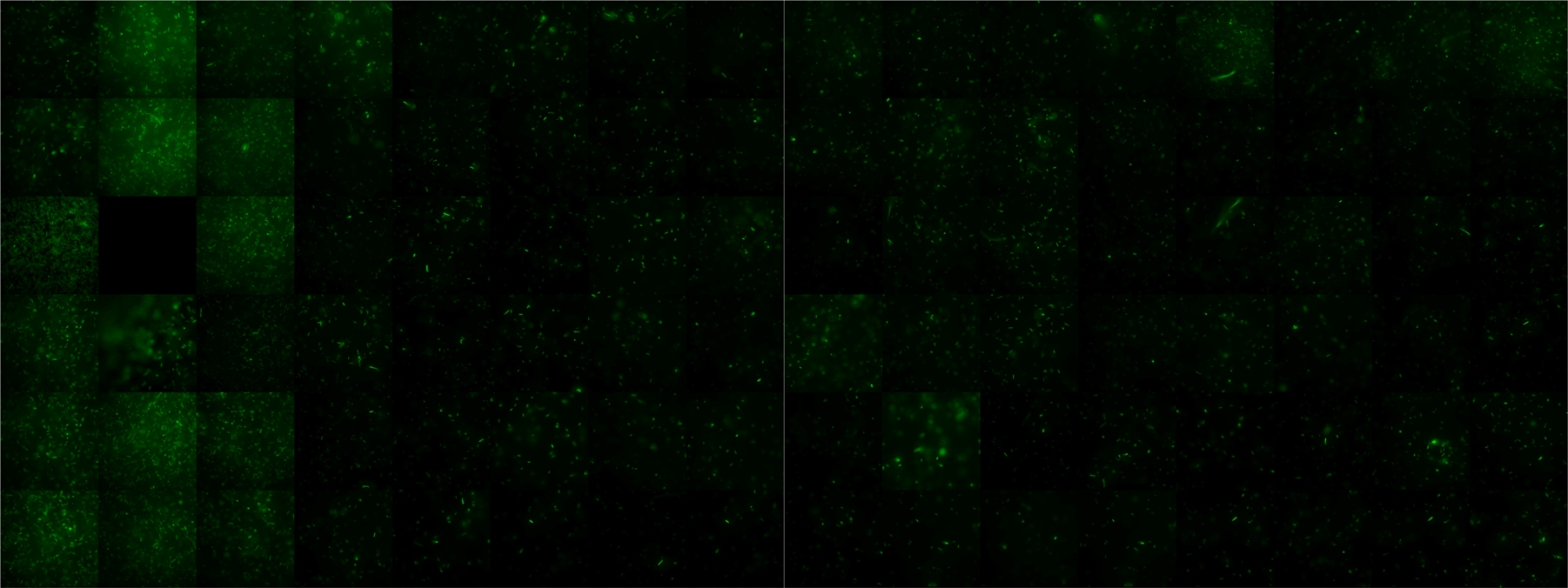
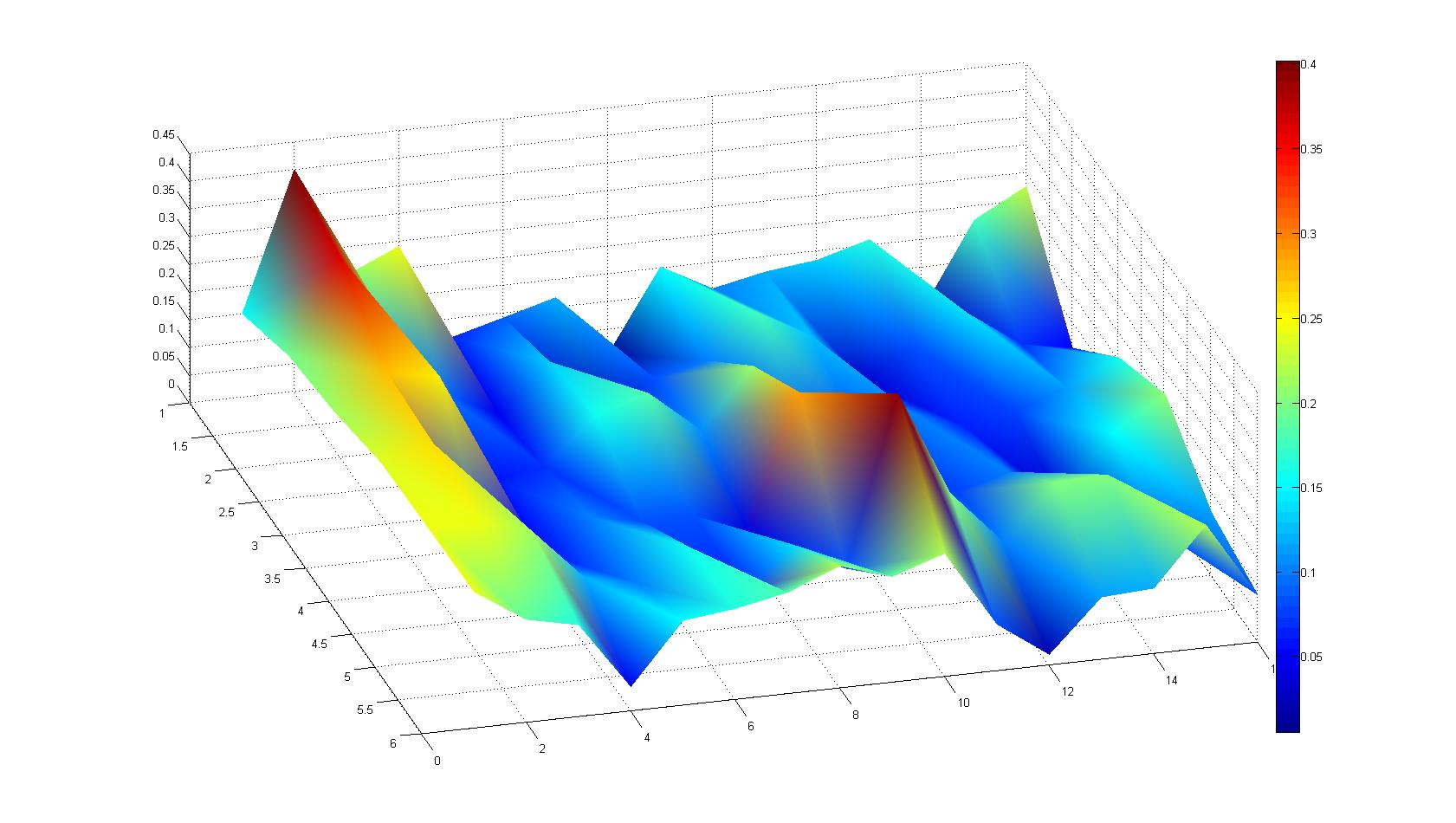
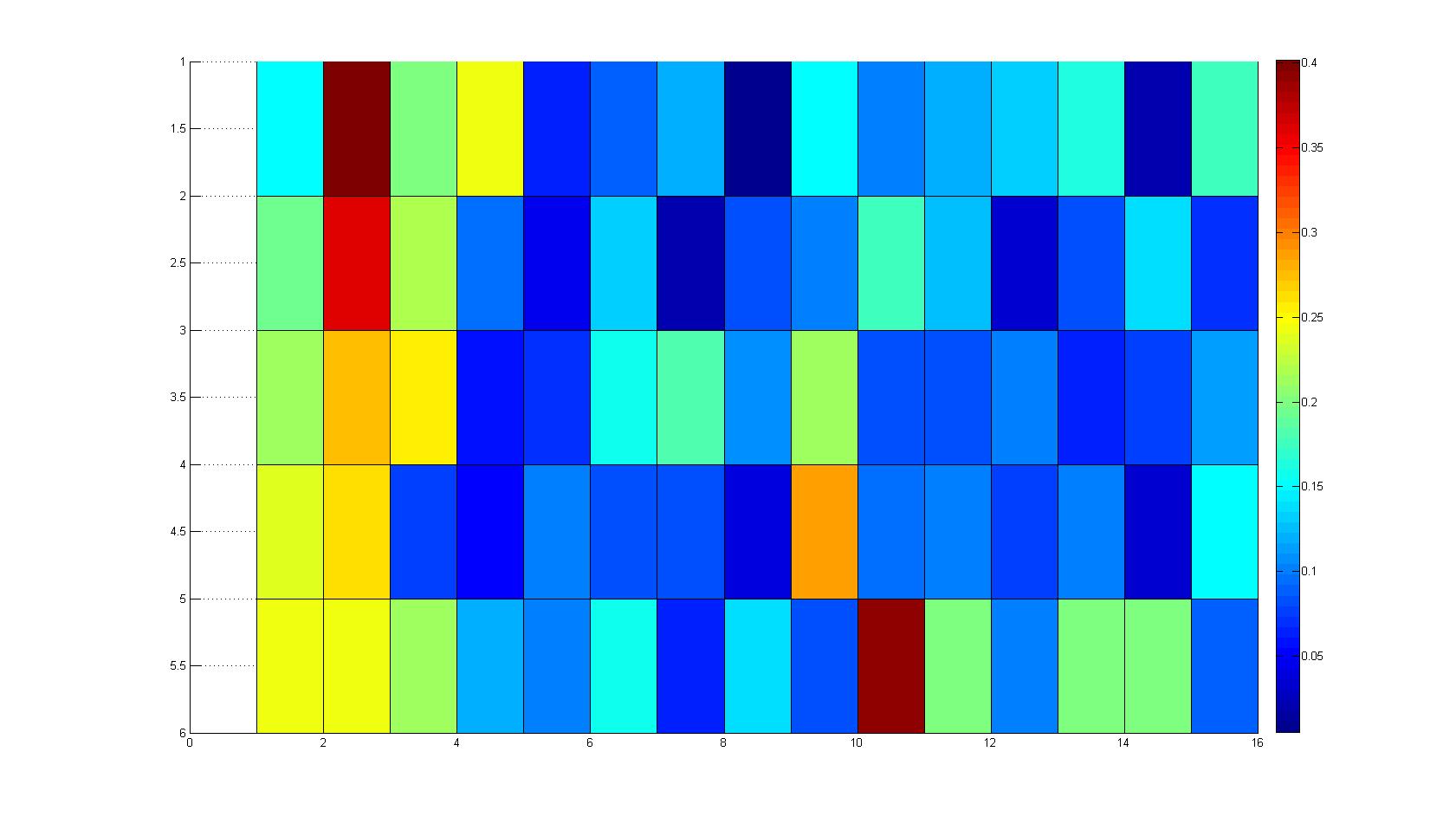
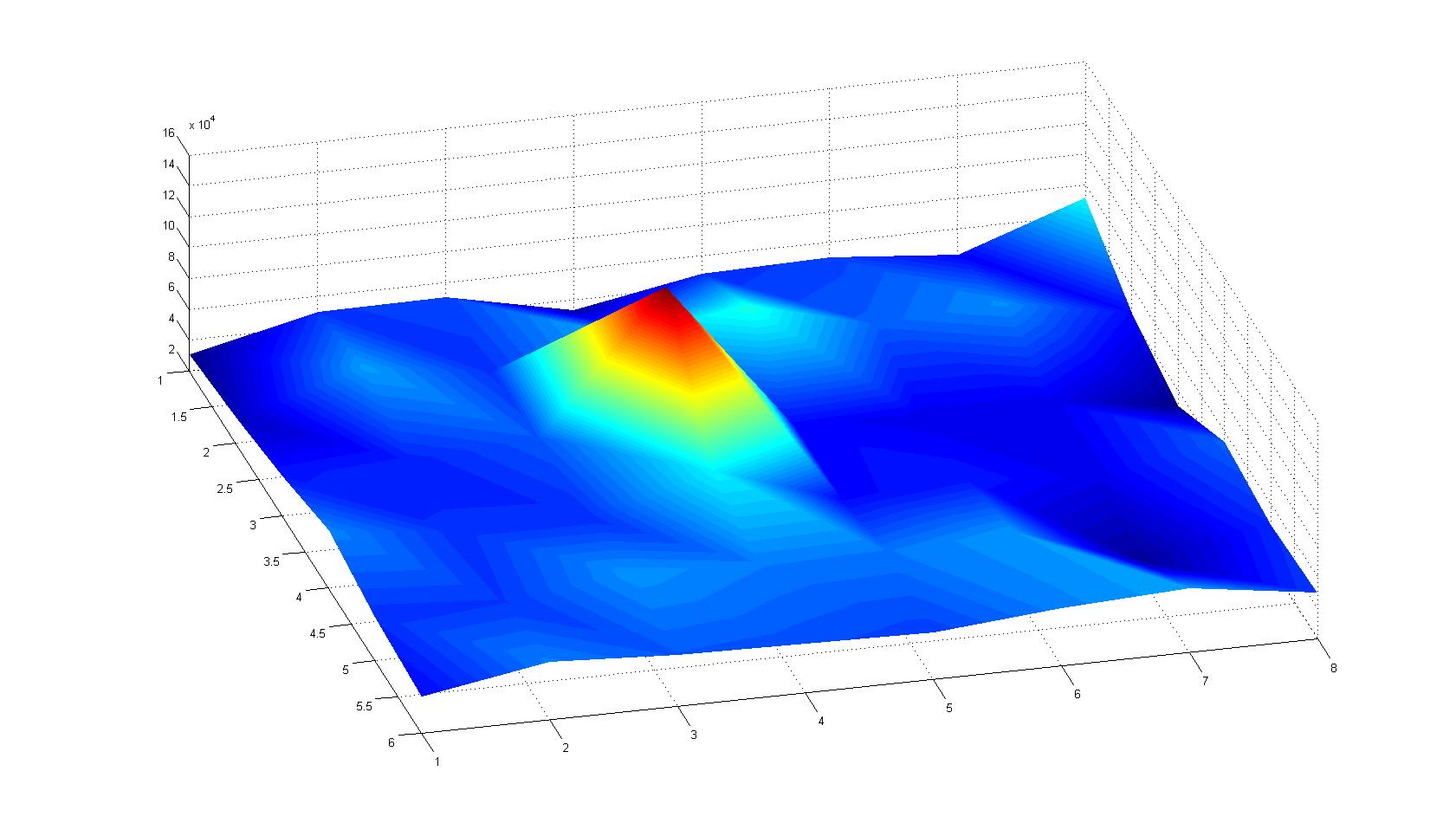
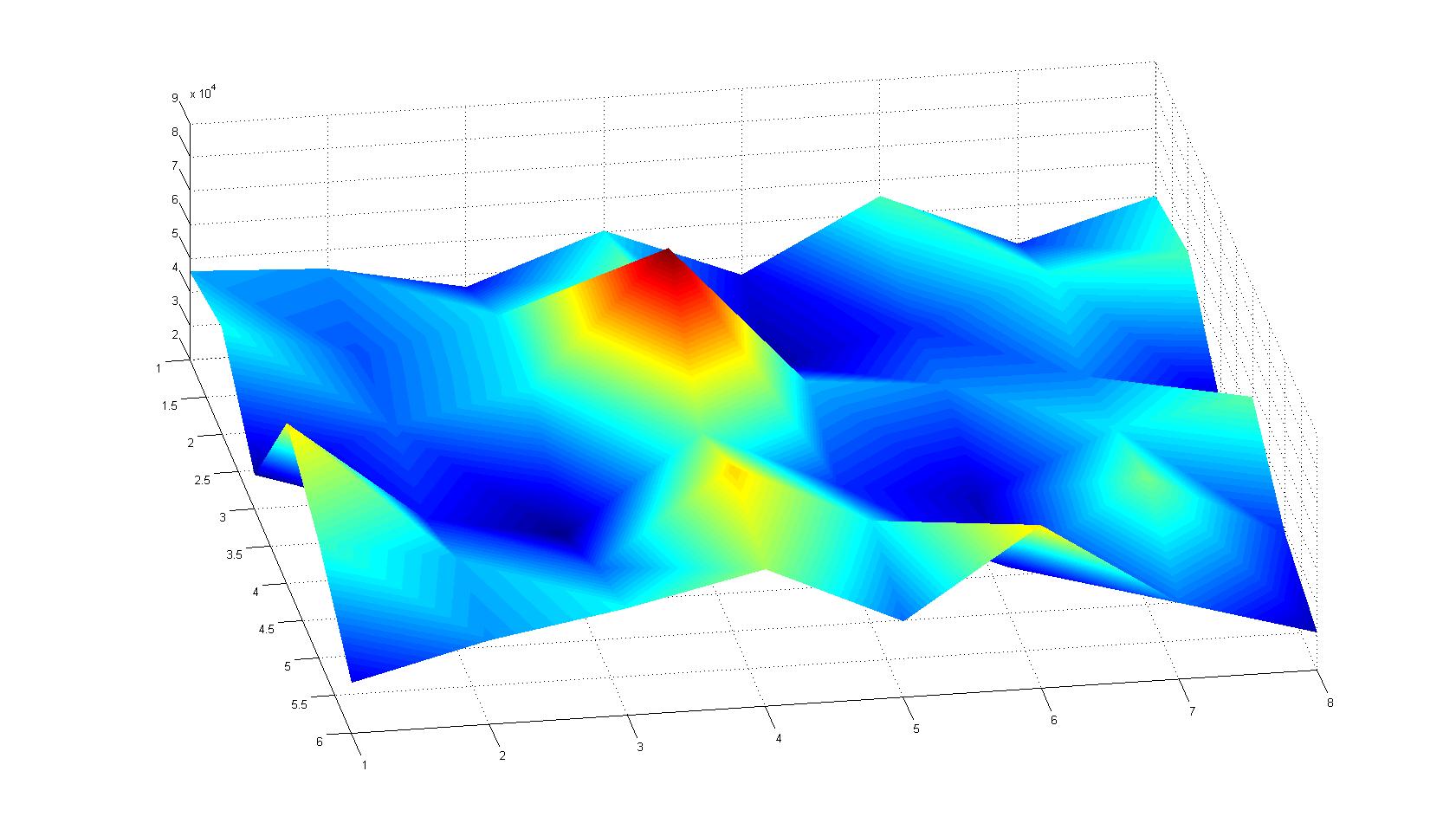
A scan of the bacterial density in a soft agar chamber (visit Notebook - Visualization for more technical information). For a proof of principle the chemotactic E. coli strain HCB33 is applied in the middle of the chamber. On the left side of the chamber is a 2% agar plug containing 10% of casamino acids. After 21 hours the bacteria accumulate around the plug, because they sense the established gradient of amino acids. Calculation of the mean of each image and subtraction of the background leads to a quantitative 2D representation of the bacterial density landscape. This data can be compared to the 2D chemotactic modeling results based on a system of partial differential equations (visit Modeling - Chemotaxis/Colicins).
Remark: The scans are taken from two different experiments with equal settings due to the difficult adjustment of a constant focus.
After 3 hours
| |
After 21 hours
[back]
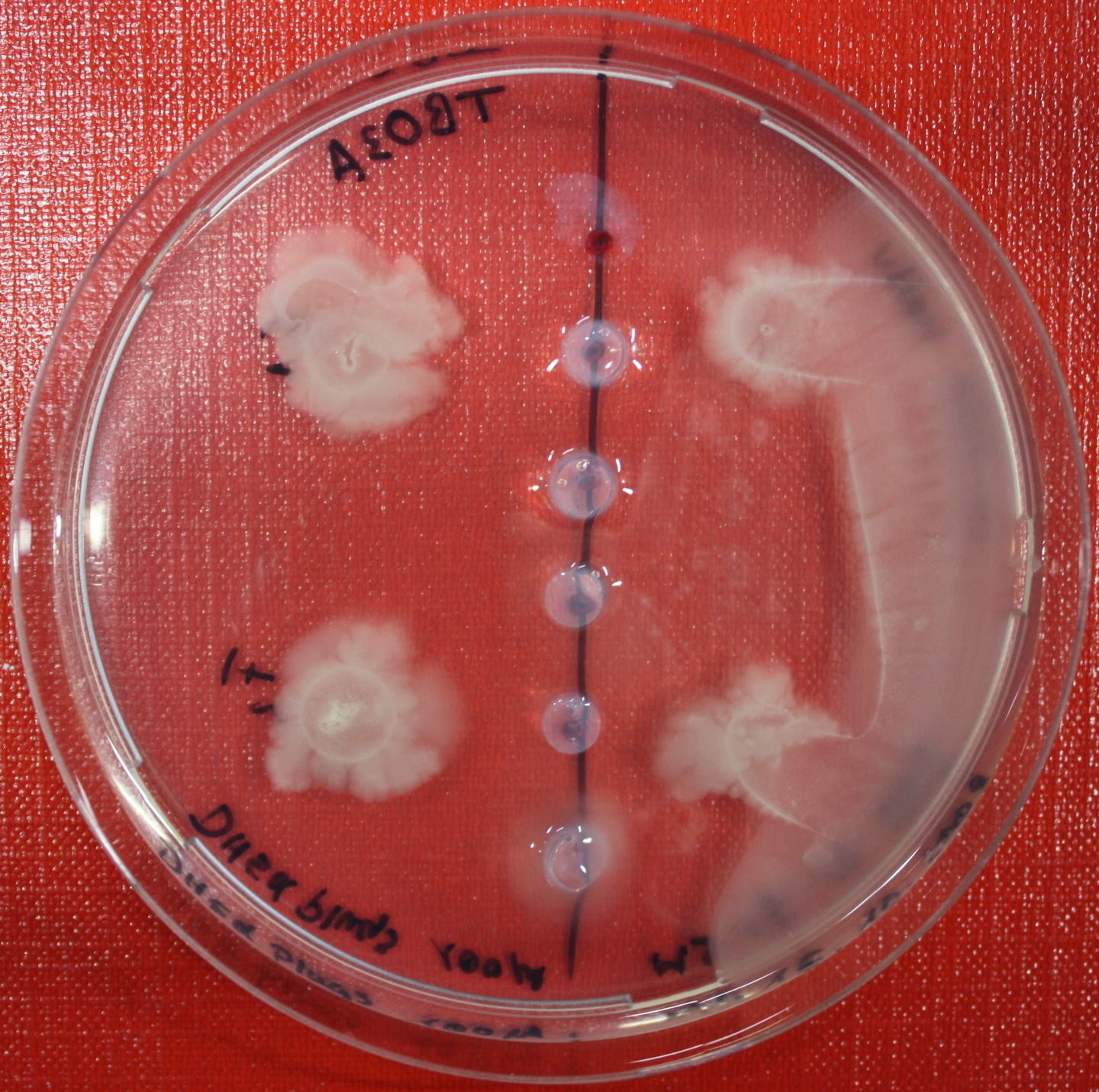
Sensing test of Fusion Receptor with 2D Microscopy
Sensitivity of Fusion constructs to AI-2 is analyzed in fluorescent UU1250 and HCB33 strains. Images are taken on a wide-field Delta-Vision microscope in LabTek chambers with TB medium, Tethering Buffer or minimal media, all containing 0.2% agar (2 ml). As attractant either 3 µl of AI-2 producing cells (OD=0.4, induced with100 nM IPTG), or 10 µl supernatant from these cells are spotted at the left side of the chamber. A gradient is established during incubation for 12-16 hours at 4 °C. Afterwards either fusion receptor expressing cells (0.01% arabinose induced overnight) or non-chemotactic reference strains are spotted at the center of the chamber. The chamber is then incubated for several hours at 30 °C.
After 0 hours
After 3 hours
[back]
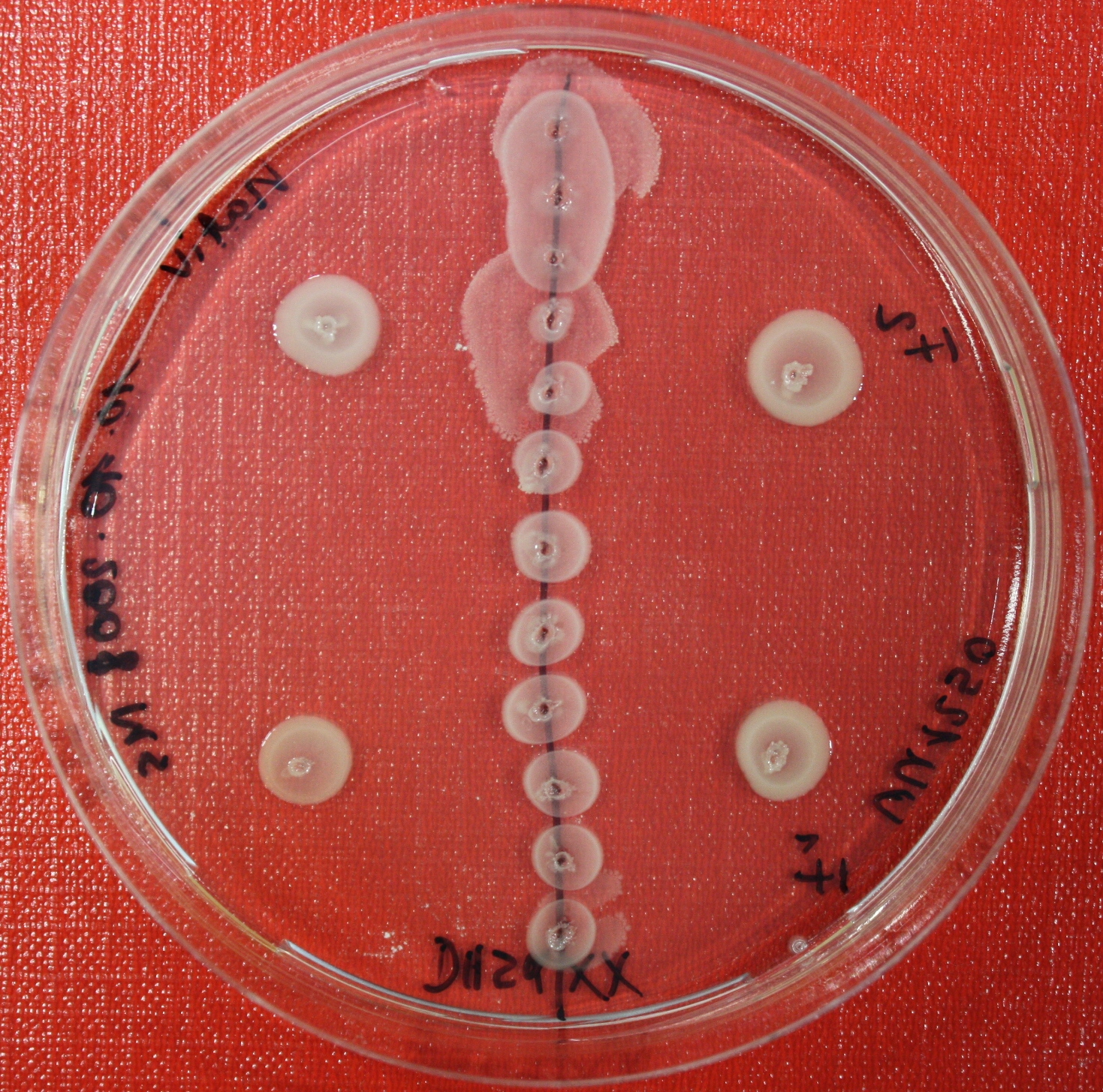
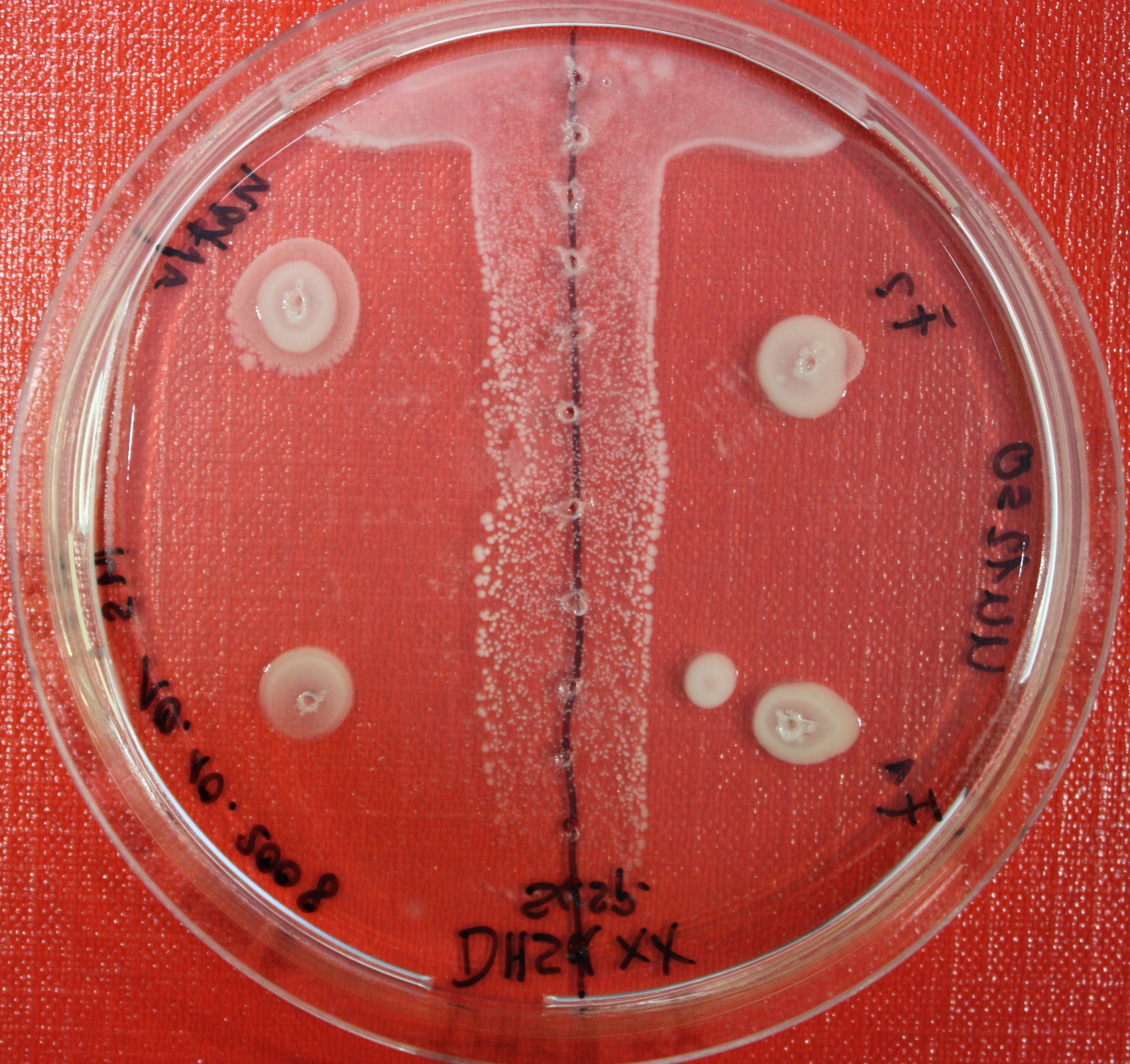
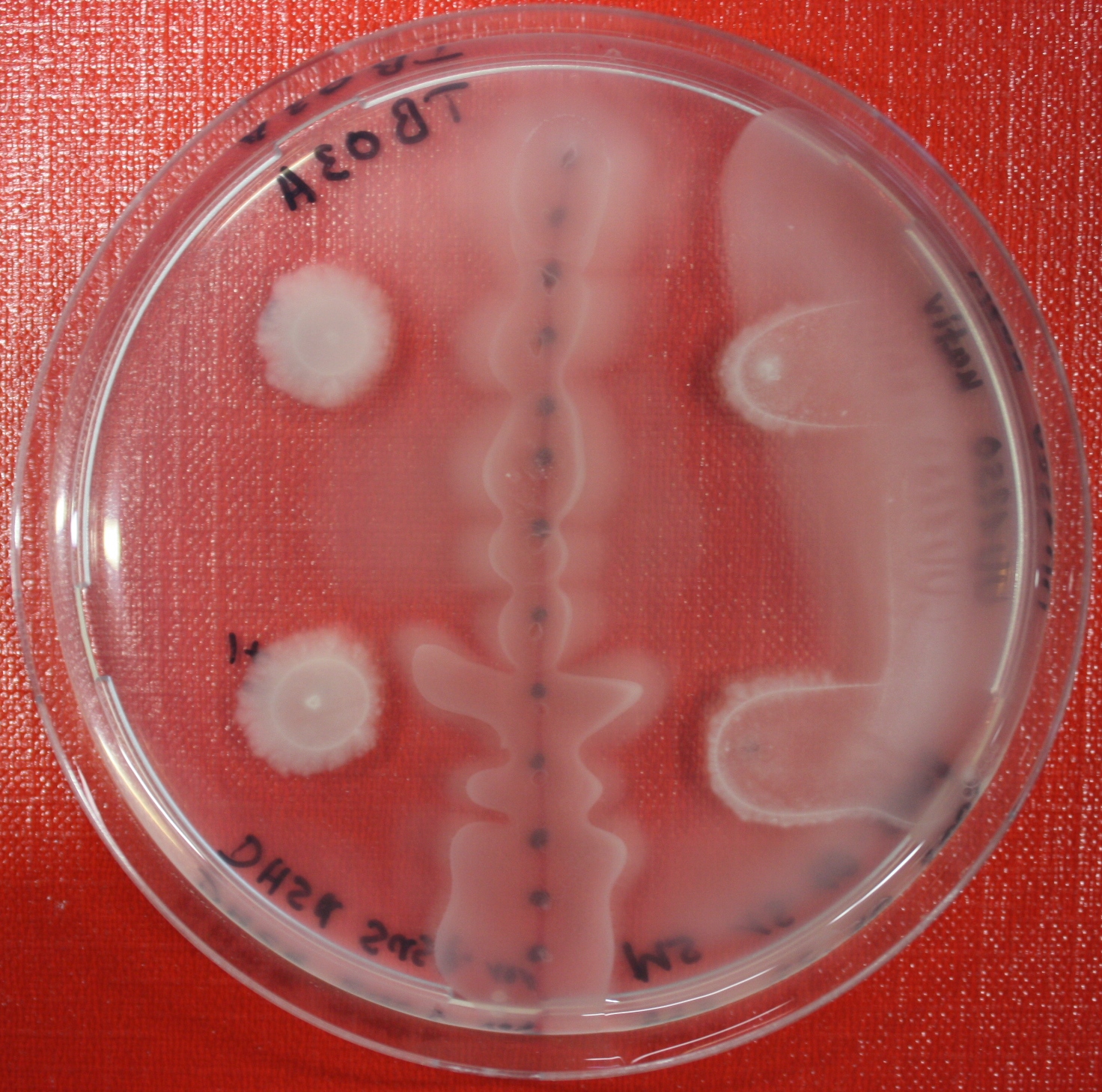
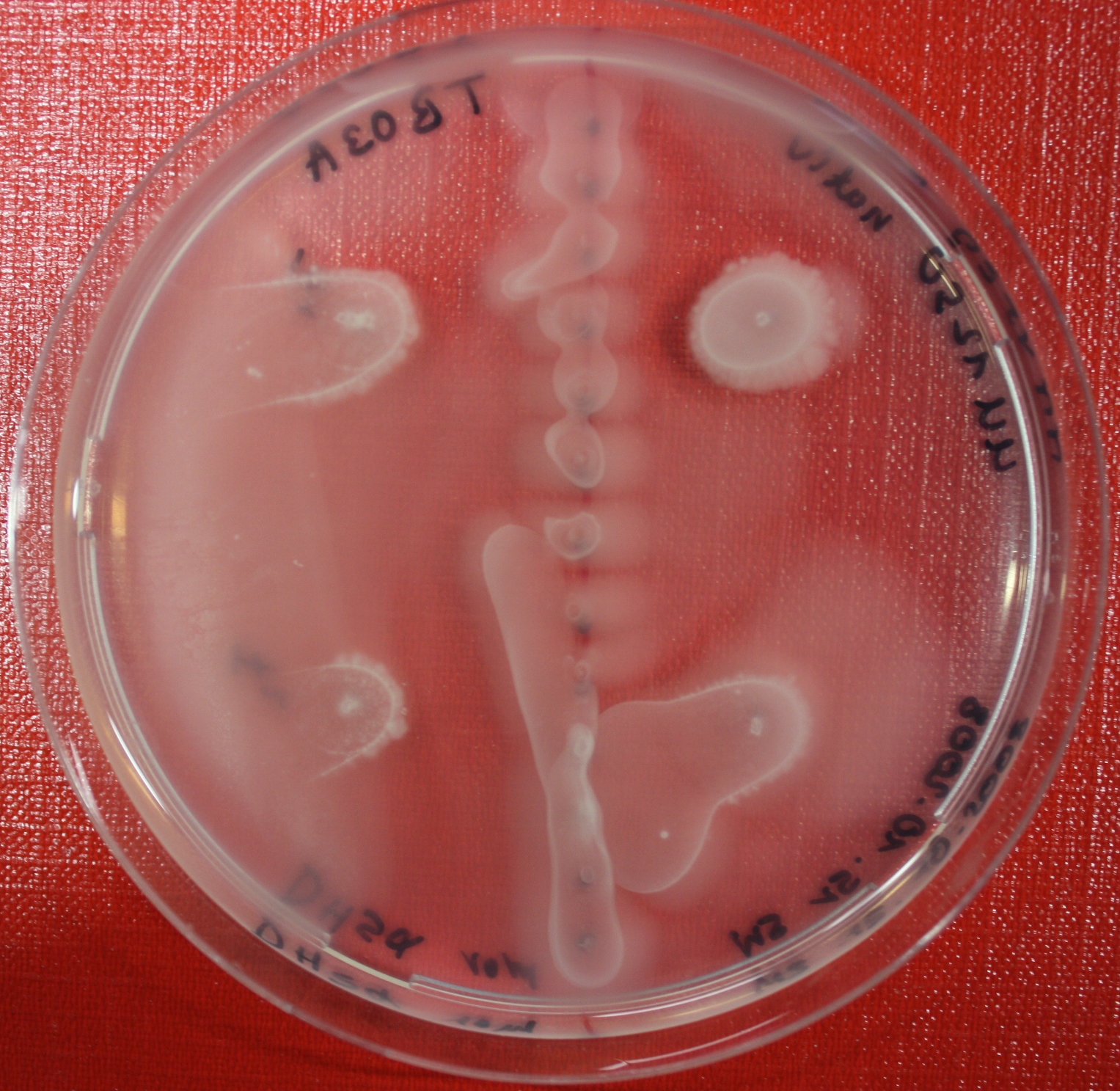
Sensing test of Fusion Receptor with Swarm Plates
Swarm assays are done to test the sensitivity of the fusion receptor to AI-2. Two different strains are used: UU1250 and HCB33. The assays are conducted in TB medium for UU1250 and TB or minimal medium for HCB33, both containing 0.3% agar. As attractant either 3 µl of AI-2 producing cells (OD=0.4), induced with 100 nM IPTG, or 10 µl supernatant from those cells are spotted 13 times on the plate to create a center line. The gradient of AI-2 is established within 12-16 hours incubation at 4 °C. The fusion receptor expressing cells (induced with 0.01% arabinose overnight) as well as non-chemotactic reference strains were spotted at a distance of 3-3.5 cm of the attractant line. Swarm plates were incubated at 30 °C and pictures taken after 24, 48 and 72 hours.
[back]
 "
"