Team:Peking University/Project
From 2008.igem.org
(→Project Background) |
(→Project Background) |
||
| Line 44: | Line 44: | ||
Inspired by this work, we hope to engineer yeast to make an in vivo evolution machine. Compared with B cell, yeast is cheap, robust and reproduces at an amazing speed, as is desired by practical applications. Moreover, yeast is a simple eukaryotic system with similar basic architecture of mammal cells, which render us to perform in vivo assay of evolved proteins for eukaryotic function without any extraction and purification. Our system has a variety of applications in academic and industrial context, theoretically almost any inter-molecular interaction that involves proteins and RNAs. In particular, we expect to use it to screen for drug target, for example, G protein coupled receptors (GPCR) which make up more than one fourth of drug targets. | Inspired by this work, we hope to engineer yeast to make an in vivo evolution machine. Compared with B cell, yeast is cheap, robust and reproduces at an amazing speed, as is desired by practical applications. Moreover, yeast is a simple eukaryotic system with similar basic architecture of mammal cells, which render us to perform in vivo assay of evolved proteins for eukaryotic function without any extraction and purification. Our system has a variety of applications in academic and industrial context, theoretically almost any inter-molecular interaction that involves proteins and RNAs. In particular, we expect to use it to screen for drug target, for example, G protein coupled receptors (GPCR) which make up more than one fourth of drug targets. | ||
| + | |||
| + | {| | ||
| + | [[Image: Background3.jpg|350px]]||[[Image: Background4.jpg|350px]] | ||
| + | |} | ||
== '''Project Details''' == | == '''Project Details''' == | ||
Revision as of 19:34, 29 October 2008

| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Misc&Fun | [http://www.openwetware.org/wiki/IGEM:Peking_University/2008 Our OWW] | [http://www.openwetware.org/wiki/IGEM:Peking_University/2008/Notebook Notebook] |
|---|
Contents |
Project Abstract
A Genetic Circuit for Directed Evolution in vivo
Directed evolution is a powerful tool for answering scientific questions or constructing novel biological systems. Here we present a simple genetic circuit for in vivo directed evolution which comprises minimal elements for random mutation and artificial selection. We engineer yeast to generate the DNA mutator hAID, an essential protein in adaptive immunity, and target it specifically to a gene of interest. The target gene will be mutated and consequently promptly evolves. By linking the expression of hAID repressor LacI and favorite gene functionality, the mutation rate inversely correlates between the functionality of the desired gene and hAID. This circuit may be adopted for in vivo evolution in eukaryotic system on genetically encoded targets. It has a variety of potential applications in academic and industrial contexts, theoretically most inter-molecular interaction that involves proteins and RNAs.
Genetic Circuit
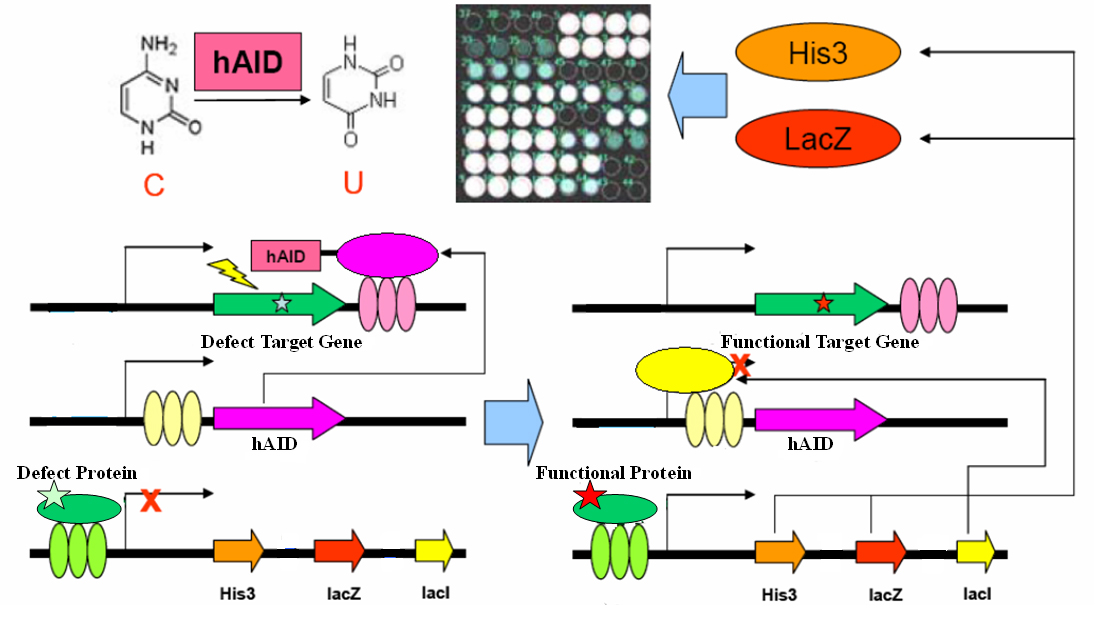
|
Project Background
Evolution has played a critical role in generating diverse organisms and proteins whose functions are perfectly coordinated with one another. So far human efforts for designing biological systems are paled by the masterpieces of nature. However, natural evolution is generally so slow that it typically takes millions of years to evolves new proteins. Therefore scientists have developed several strategies for protein evolution in vitro, such as error-prone PCR mutagenesis, mRNA display, DNA shuffling and phage display, etc. These strategies successfully reduce the cost of time by a significant degree. Nevertheless, high-throughput screening still requires a great deal of man power as well as time. Besides, the separation of evolution and screening add complexity to experimental procedures.
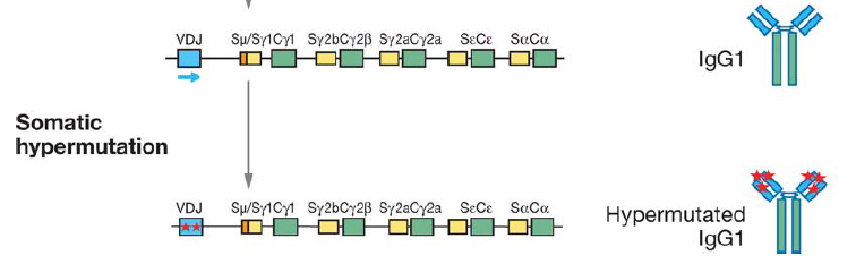
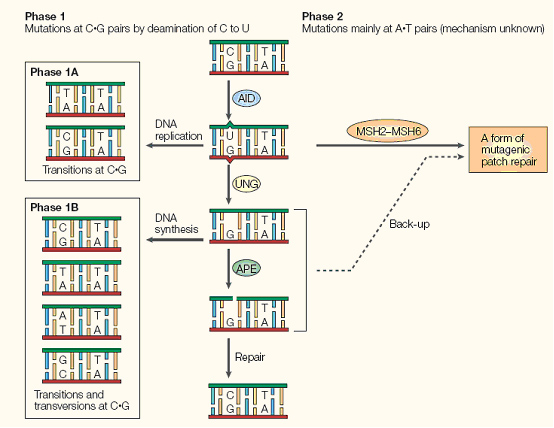
However, we overlooked a mechanism natural has adopted for rapid evolution of proteins: adaptive immunity that generates a huge pool of antibodies.It is well-known that B-cells are capable of producing a large pool of new antibodies upon antigen stimuli, which is a most important protective mechanism for animals. People have been curious about the molecular mechanism of antibody generation. Recent works have revealed that an enzyme: Activation-Induced (Cytidine) Deaminase (AID) serves as an essential protein in three processes for antibody diversity, namely somatic hypermutation (SHM), class switch recombination (CSR) and gene conversion. Briefly speaking, AID converts cytidine to uracil by oxidizing the amino group to carbonyl group, resulting in mismatch of Watson-Crick base pair. Then DNA lesion repair pathways (base excision repair, BER; mismatch repair, MMR)are employed to bring DNA back to normal. In this process, the coding sequence for the hypervariable region of immunoglobulin is changed at rapid pace and hence generates diverse antibodies. AID target must be specific in order to avoid the risk of B cell lymphocytoma. AID is proved to be functional in several other cells. In 2005, Youri Pavlov expressed human Activation-Induced Deaminase (hAID) in Saccharomyces cerevisiae (budding yeast) and found that AID would cause genomic mutation at a rate of 10-6.
(Neuberger, M.S., et al., Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat Rev Immunol, 2005. 5(2): p. 171-8.[http://www.ncbi.nlm.nih.gov/pubmed/15688043?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVMedline PubMed])
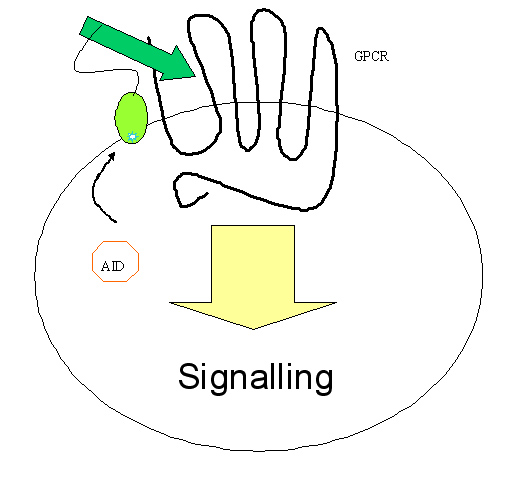
Inspired by this work, we hope to engineer yeast to make an in vivo evolution machine. Compared with B cell, yeast is cheap, robust and reproduces at an amazing speed, as is desired by practical applications. Moreover, yeast is a simple eukaryotic system with similar basic architecture of mammal cells, which render us to perform in vivo assay of evolved proteins for eukaryotic function without any extraction and purification. Our system has a variety of applications in academic and industrial context, theoretically almost any inter-molecular interaction that involves proteins and RNAs. In particular, we expect to use it to screen for drug target, for example, G protein coupled receptors (GPCR) which make up more than one fourth of drug targets.
Project Details
Project Proposal
Evolution could be dissociated into two parallel processes: random mutation, and natural selection. Whilst its role in life history is well understood and accepted, evolution is also evident across individual life process. In particular, adaptive immunity system adopts evolution strategy to produce antibodies for novel antigens. Artificial evolution method could be a powerful tool for answering scientific questions or engineering novel biological systems. Via systems biology approach, here we present a simple genetic circuit consisting functional elements for random mutation and artificial selection. This circuit may perform in vivo evolution on virtually any genetically encoded targets, with potential applications in academic and industrial contexts.
The gene encoding the core element in adaptive immunity, hAID, is fused with the LexA-DBD domain with flexible linker. The fusion gene hAID-LexA is inserted into a yeast ESC expression cassette at 3’ of tandem LacO elements. The target gene (your favorite gene, yfg) is inserted into a yeast ESC expression cassette, with its 3’UTR containing tandem LexO elements. An inducible promoter response to yfg activity drives expression of His3, LacZ and LacI in cistrone spanned by IRES. These three plasmids together form a genetic circuit for in vivo evolution.
For clearness, here we present one very simple example in yeast one hybrid: a mutated Gal4 gene is inserted in the target cassette. The selector cassette contains UAS to drive expression of His3, LacZ and LacI. All three plasmids are transfected or knock-in-ed into gal4-, his3- yeast strain with proper selection tags. Initially, we culture the yeast in complete YPD medium. Defect Gal4 product cannot bind to UAS, hence hAID-LexA is constitutively expressed, recruited to LexO sites of the target, and mutates the Gal4 conding sequence. Once Gal4 mutation is reversed, it binds to UAS to drive LacI expression, which represses hAID-LexA. We plate the yeast into Trp-, His-, Leu-, 3AT+, XGal+ plate and select for the large, blue colonies. Sequencing the colonies then gives us the activated Gal4 gene sequences.
The power of this system could be best revealed in the above case: since selection is not required for initiation and attenuation of mutagenesis, there is virtually no need to perform selective pressure titration.
The system could be readily adopted to in vivo evolution of any kind of protein-protein and protein-nucleic acid interaction: for this we simply adopt a yeast two hybrid-like approach, evolving a functional protein linked to Gal4 AD which could bind to the "bait" protein linked to Gal4 DBD. We could also use the yeast three hybrid system to study RNA-protein interaction, or use yeast one hybrid to evolve specific protein that binds to given promotor.
Yeast shows several advantages in studying eukaryotic process, for it folds and modifies the eukaryotic protein accurately, whilst bacteria does not. Another advantage of yeast is that cell wall blocks intracellular communication between transmembrane proteins, and the selection happens completely within the cell, therefore blocking possible false positives. We plan to screen for extracellular activator of certain eukaryotic transmembrane protein, for example, novel peptidergic ligand for GPCR and antibody-antigen interaction.
We have been working on further improvements of the mutation system, by improving the enzyme to archieve directed, "hotspot-less", and evenly distributed mutagenesis, by utilizing the overwhelming power of yeast genetics, and by harnessing novel protein chemistry.
//Last modified: ZY 20080704
The Experiments
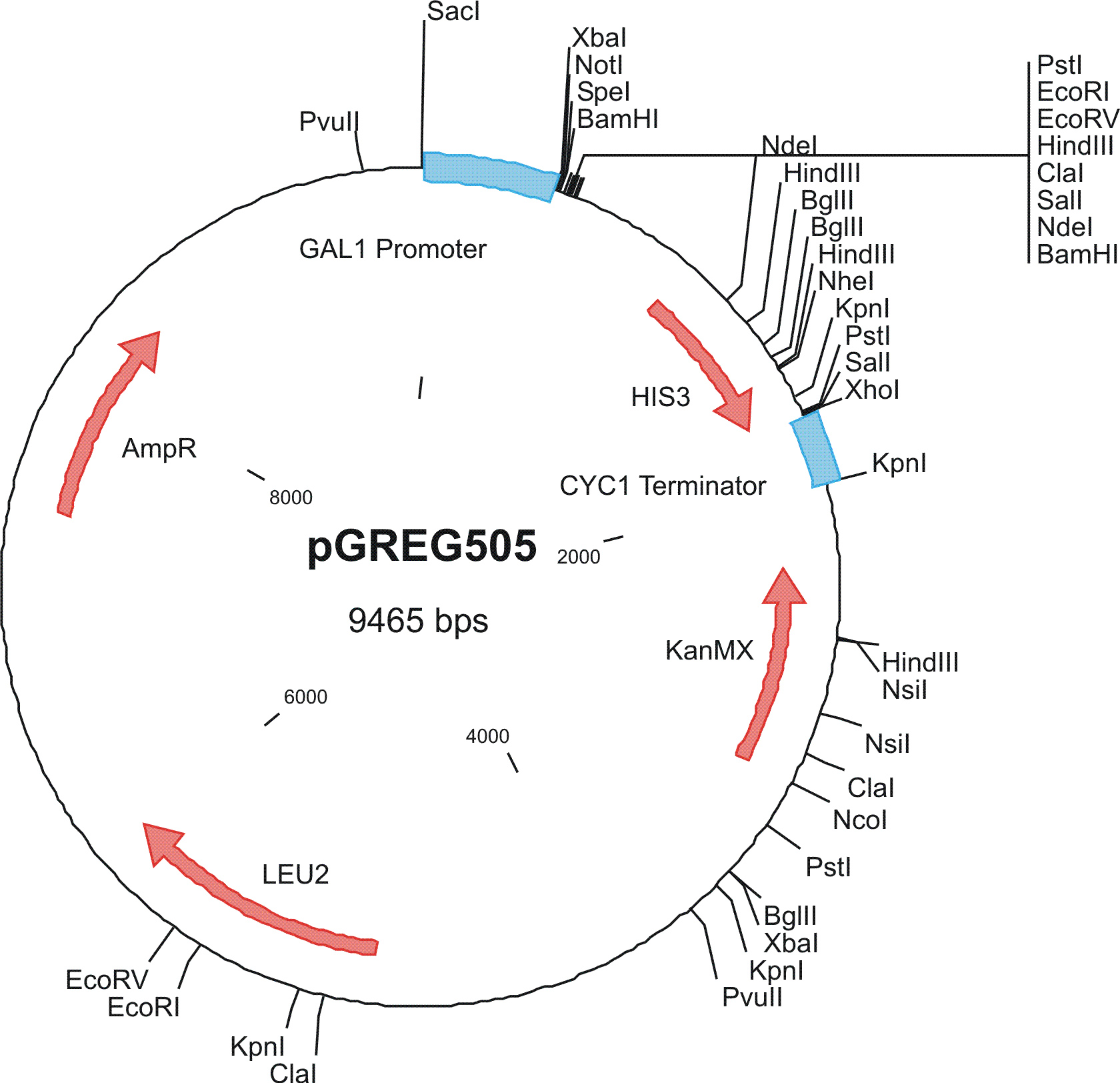
To construct the genetic circuits above, we have used the pGREG series of vectors which were hosted in budding yeast Saccharomyces cerevisiae AH109. pGREG503, the expression plasmid with HIS3 auxotroph resistance was used to construct the UAS-lacI component. LacI was under the galactose inducible promoter GAL1. The promoters of pGREG504 and pGREG505 were replaced by pADH-lac and pACT respectively. pADH-lac is pADH based promoter which is sensitive to lacI, genes under this promoter will be inhibited by lacI. pACT is a constitutively expressing promoter derived from the promoter of actin in budding yeast. The construct hAID-linkers-lexA was under the pADH-lac promoter and the GAL4-lexO construct was engineered under the pACT promoter. Notably, in the genome of the yeast strain AH109, it has GAL1-HIS3 and GAL1-lacZ constructs. PGREG504-hAID-linkers-lexA DBD was constructed via the following Figure.
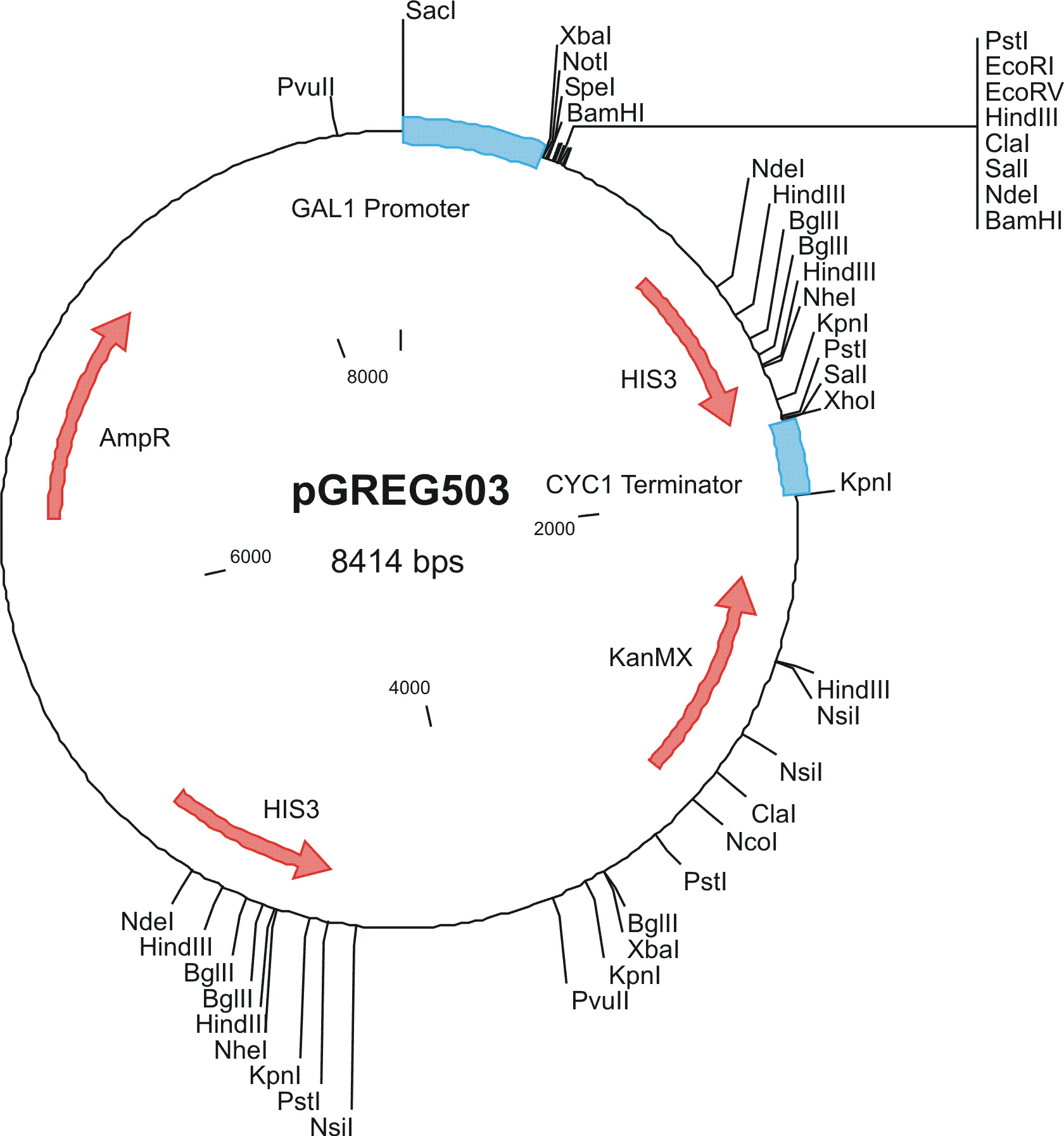
| 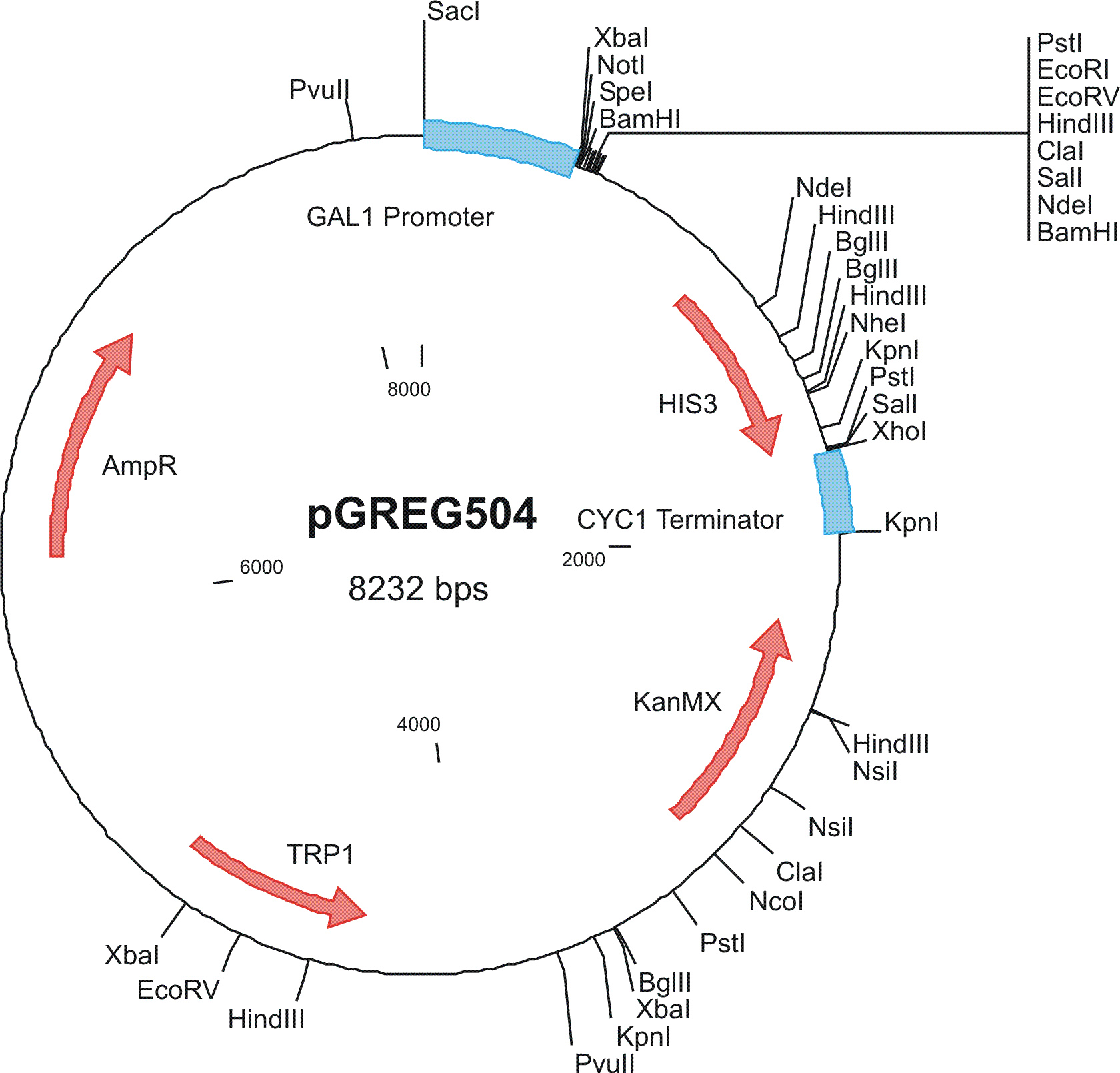
| 
|
To construct plasmid on pGREG503, we have used the versatile PCR-based homologous recombination system: the PCR product of lacI was directly co-transformed with pGREG503 and it was automatically recombined to the site flanking the 3’ of the promoter. Other constructs were accomplished with standard protocols.
 "
"