DNA-Origami
From 2008.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
style="font-weight: bold;">Cell culture</span><br> | style="font-weight: bold;">Cell culture</span><br> | ||
</h4> | </h4> | ||
| - | 50 ml DYT-Medium, | + | 50 ml DYT-Medium, 50 µl tetracycline (TET; 25 mg/ml) and ER2738-cells |
were shaken over night at 37°C. The overnight culture was diluted with | were shaken over night at 37°C. The overnight culture was diluted with | ||
| - | DYT to OD600=0.1 and shaken at 37°C until the culture | + | DYT to OD600=0.1 and shaken at 37°C until the culture reached an OD600 |
around 0.4. Each 50 ml of cell culture were inoculated with 5 µl | around 0.4. Each 50 ml of cell culture were inoculated with 5 µl | ||
M13mp18 phage and shaken for 4 h at 37°C.<br> | M13mp18 phage and shaken for 4 h at 37°C.<br> | ||
| Line 28: | Line 28: | ||
transferred in a falcon tube. The phages stay in the | transferred in a falcon tube. The phages stay in the | ||
supernatant therefore the supernatant was carefully decanted to the | supernatant therefore the supernatant was carefully decanted to the | ||
| - | PEG/NaCl and mixed gently by inverting the tube. The mixture ( | + | PEG/NaCl and mixed gently by inverting the tube. The mixture (solution |
1) was left overnight at 4°C. <br> | 1) was left overnight at 4°C. <br> | ||
Solution 1 was centrifuged at 5000 g for 20 min. Because the phage stay | Solution 1 was centrifuged at 5000 g for 20 min. Because the phage stay | ||
in the pellet, the supernatant was removed and the pellet was | in the pellet, the supernatant was removed and the pellet was | ||
| - | resuspended in 2 ml TBS-Buffer ( | + | resuspended in 2 ml TBS-Buffer (solution 2). Solution 2 was put in a |
| - | 1 | + | 1.5 ml Eppendorf tube and centrifuged (13200 rpm, 10 min). After the |
centrifugation the phages stay in the supernatant.<br> | centrifugation the phages stay in the supernatant.<br> | ||
<span style="font-weight: bold;">Second precipitation</span><br> | <span style="font-weight: bold;">Second precipitation</span><br> | ||
170 µl PEG/NaCl(~ 1/6 volume of supernatant) were put in a Eppendorf | 170 µl PEG/NaCl(~ 1/6 volume of supernatant) were put in a Eppendorf | ||
tube. Supernatant was carefully decanted to the PEG/NaCl and mixed | tube. Supernatant was carefully decanted to the PEG/NaCl and mixed | ||
| - | gently by inverting the tube. The mixture ( | + | gently by inverting the tube. The mixture (solution 3) was left for 1 h |
on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | ||
<h4>Define phage titers</h4> | <h4>Define phage titers</h4> | ||
| - | The absorption of | + | The absorption of solution 3 was measured on a Jasco V-550 UV/VIS |
spectrometer at 269 nm. <br> | spectrometer at 269 nm. <br> | ||
Phage titer was calculated as follows:<br> | Phage titer was calculated as follows:<br> | ||
| Line 62: | Line 62: | ||
[[Image:TeamFreiburg2008_Tabellen-Origamipipettier-Schema2.jpg|800 px]]<br> | [[Image:TeamFreiburg2008_Tabellen-Origamipipettier-Schema2.jpg|800 px]]<br> | ||
Various samples were | Various samples were | ||
| - | produced. For the sample with a ratio of 1:20 (DNA:Oligo) we used 4 nM | + | produced. For the sample with a ratio of 1:20 (DNA:Oligo), we used 4 nM |
| - | DNA and 80 nM of oligos. The Origami were produced in | + | DNA and 80 nM of oligos. The Origami were produced in an eppendorf |
| - | Mastercycler personal. | + | Mastercycler personal. They were heated up to 95°C for 7 min |
and slowly cooled down (0.3°C/s) to 20°C.<br> | and slowly cooled down (0.3°C/s) to 20°C.<br> | ||
Different sample were made:<br> | Different sample were made:<br> | ||
| Line 75: | Line 75: | ||
<h4><span style="font-weight: bold;"></span>Purification of the DNA-Origamis<br> | <h4><span style="font-weight: bold;"></span>Purification of the DNA-Origamis<br> | ||
</h4> | </h4> | ||
| - | To purify the DNA-Origamis from the unbound DNA-oligos we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on | + | To purify the DNA-Origamis from the unbound DNA-oligos, we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on top in 1.5 ml Eppendorf tubes. To clean the filter of remaining glycerol, 450 µl TAE/MgAcetat (12.5 mM; 1x filtered) was put on top of the filter and centrifuged for 15 min at 1000 g. After removing the filtrate, 400 µl TEA/MgAcetat (12.5 mM;1x filtered) and 45 µl DNA-origami were put on top of the filter and again centrifuged for 15 min at 1000 g. The filtrate was removed again. All unbound DNA-oligos were washed off by putting 400 µl TEA/MgAcetat (12.5 mM; 1x filtered) on top of the filter. The sample was centrifuged for 15 min at 1000 g. To release the DNA-origamis of the filter, 100 µl TAE/MgAcetat (12.5 mM;1x filtered) was put on top of the filter, and the filter was left at room temperature for at least 2 min. The filter shouldn´t run dry. The Montage® PCR Centrifugal Filter Devices were put upside down (the purple side has to be on the bottom) in one of the special Invert Spin tubes form Millipore and centrifuged for 3 min at 1000 g. |
| - | The Origami were kept in different buffers. For this TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> | + | The Origami were kept in different buffers. For this, TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> |
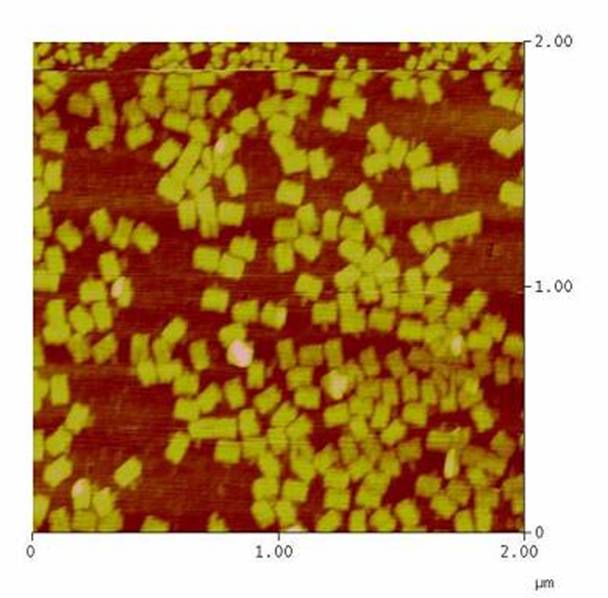
<h4><span style="font-weight: bold;"></span>Atomic force microscopy to prove if the origami stable | <h4><span style="font-weight: bold;"></span>Atomic force microscopy to prove if the origami stable | ||
<br> | <br> | ||
</h4> | </h4> | ||
To see if the Origami were formed well and stable in the different buffers an atomic force microscope (AFM) was used. The measurement itself was done in air (not in the buffer). | To see if the Origami were formed well and stable in the different buffers an atomic force microscope (AFM) was used. The measurement itself was done in air (not in the buffer). | ||
| - | The DNA-Origami were absorbed to freshly cleaved mica. Therefore the mica was cut into 6 mm pieces and affixed to the metal panes we used for the measurement. To get | + | The DNA-Origami were absorbed to freshly cleaved mica. Therefore the mica was cut into 6 mm pieces and affixed to the metal panes we used for the measurement. To get an atomically clean surface, an adhesive tape was used to remove the topmost mica layers. After this the sample could be put on. First 2-10 µl of the sample were put on the mica and then quickly diluted with water (just as much that the mica was covered with fluid). The sample was incubated for about 5 min and then the mica was blown dry with a stream of nitrogen. Then the sample could be measured. |
The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | ||
<h2>Results and Discussion</h2> | <h2>Results and Discussion</h2> | ||
Revision as of 22:27, 29 October 2008
 "
"