|
Overview
In order to approach our goal of creating an E. coli strain carrying a minimal genome, there are three main problems that have to be overcome:
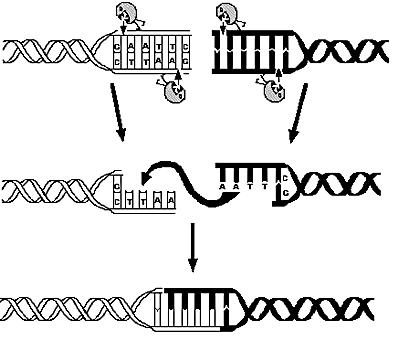
Genome reduction: show that in vivo restriction and religation is possible
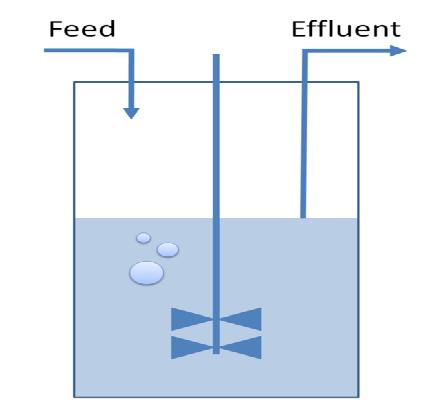
Chemostat selection: introduce a limitation that confers a growth advantage to organisms with smaller genomes
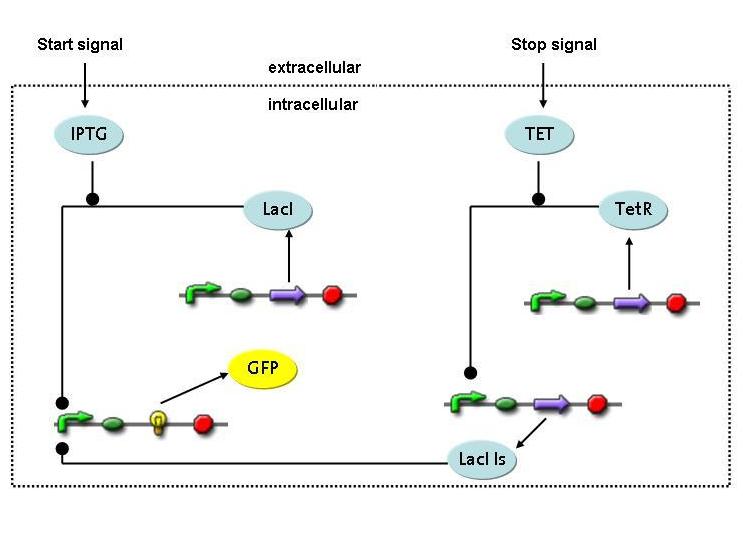
Switch circuit: design a biobrick that provides for short-term synthesis of the desired gene products
To prove that in vivo restriction and religation is possible is fundamental to our project which relies on short-term expression of a restriction enzyme and a ligase. While the restriction enzyme will randomly cut DNA, the simultaneous or shortly delayed synthesis of the ligase should religate the DNA. If the DNA is cut at several sites, religation will lead to exclusion of chromosomal fragments in a random manner.
In the continuous culture of a chemostat, those organisms with the highest rate of proliferation will overgrow those with a smaller growth rate. In order to bypass the need of selecting for those E. coli which have successfully reduced their genomes by massive screening of thousands of clones, we need to introduce a constraint that confers a growth advantage to organisms with smaller genomes. We have chosen to introduce mutations in the nucleotide synthesis pathway to achieve this goal. This will render DNA replication the rate-limiting step of proliferation and therefore be advantageous to organisms with small genomes.
Expression of restriction enzymes that cut genomic DNA inside the cell is likely to decrease viability. Actually, the Waterloo iGEM team is using restriction enzymes to kill the cell in their project this year. Therefore, construction of a switch circuit, which allows to restrict expression of the restriction enzyme to a short period of time, is a crucial part of the project.
On the following pages, we will show a detailed description of how we are trying to achieve these three goals.
Modelling Framework
|
1) Restriction Enzymes Analysis
Questions:
- Which are the available restriction enzymes and cutting patterns?
- How is the distribution of the genes in each fragment related to the frequency of cutting?
- Is it possible to identify a restriction enzyme that optimizes the probability of reduced genome that retains vital strains?
Method:
E.Coli K12 genome was digested using 713 different restriction enzymes and, using annotation information, simple statistical analysis was applied on the calculated fragments.
Results:
The property of restriction enzymes are all related to their frequency of cutting. The mean number of genes per fragment, as well as its variance and the probability of containing essential genes, can be derived only from frequency information.
|
2) Genome Scale Analysis
Questions:
- Is it possible to slow growth of large genome strains by using a thymidine auxotrophyc strain and limiting thymidine feeding?
- What is the best restriction enzyme to be used in order to maximize genome reduction and at the same time vitality (growth rate) of thymidine auxotrophyc strains?
- What is the predicted genome reduction difference if the medium is minimal or very rich in terms of nutrients?
- Which are the predicted quantitative differences in terms of growth rate and genome size of strains on which has been applied the selection procedure?
Method:
The state-of-the-art genome scale model for E.Coli iAF1260 (1,260 genes included) was modified in order to account for thymidine auxotrophycity, thymidine uptaking limitation, genome reduction and growth on different medium. Stochastic algorithm and flux balance analysis were applied to predict growth rates.
Results:
Models show that is indeed possible to select reduced genome strains using thymidine limitation. The quantification shows that the method is at the border line with the sensitivity of chemostat machinery setup for small differencies, but is effective for big reductions (from approximately 10 Kbp on). Predictions show the possibility of reducing up to 61 % of genes for a minimal medium growing strains (corresponding to 59% of chromosome size) and 73 % of genes for rich medium growing strains (corresponding to 71% of chromosome size).
|
|
3) Chemostat Selection Method
Questions:
- What are the ideal settings of nutrient concentrations and influx in order to select reduced strains?
- Which is the sensitivity regarding growth rate selection?
- What are the timing parameters that frame the induction of two subsequent rounds of restriction enzyme expression?
Method:
A classical chemostat model using Ordinary Differential Equations was constructed and analyzed in terms of sensitivity analysis and simulation of realistic data.
Results:
|
4) Switch Circuit
Questions:
- Is it possible to design a simple but effective system for initiating and terminating restriction enzyme expression in a very tight way?
- What are the biological parameters important in such a design?
- What are the temporal constrains for the device to be used in the selection scenario?
Method:
A novel transcriptional circuit able to sense an initiating and terminating signal and to control a pulse of restriction enzyme was designed and analyzed using Ordinary Differential Equations.
Results:
The simulations show that the designed circuit is indeed able to produce a pulse-shaped response in the protein of interest which can be turned on and off by the two different inducers. Moreover the gene expression can be influenced by changing the degradation rate of the protein, for instance by tagging it, or modifying its repressors transcription rate. The pulsing frequency is mainly constrained by the time it takes for the inducers to be washed out of the medium.
|
|
 "
"