Team:University of Washington/Project
From 2008.igem.org
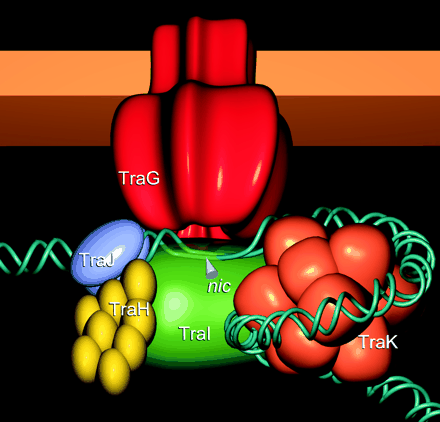
Illustration of Tra1 Conjugation Machinery
Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [http://www.ncbi.nlm.nih.gov/pubmed/11976307].
| You can write a background of your team here. Give us a background of your team, the members, etc. Or tell us more about something of your choosing. | |
|
Tell us more about your project. Give us background. Use this is the abstract of your project. Be descriptive but concise (1-2 paragraphs) | |
| Team Example 2 |
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
(Or you can choose different headings. But you must have a team page, a project page, and a notebook page.)
Contents |
Overall project
Your abstract
Project Details
Part 2
The Experiments
Part 3
Results
UW Lab Links
Protocols
BioBrick Parts Used
UW Glycerol Stocks
Welcome to The Johns Hopkins iGEM
The International Genetically Engineered Machines Competition (iGEM) is an annual intercollegiate challenge that seeks to answer the question: "Can simple biological systems be built from standard, interchangeable parts and operated in living cells? Or is biology simply too complicated to be engineered in this way?" The iGEM teams work over the summer at their respective schools, and then attend the iGEM Jamboree in November to present their work. The newly-formed iGEM team at Johns Hopkins University is composed of primarily undergraduate students with diverse majors ranging from Materials Science Engineering to Biology. While the team has graduate and faculty input, it is almost completely undergraduate run. We plan to create novel genetic parts that could be added to the existing iGEM registry of biological parts. The MIT iGEM Registry of Parts is the databank of biological standards to which iGEM teams from 2003 and onward have contributed. Currently, the databank only has 16 yeast "biobricks". These biobricks are reporters, tags, plasmids and other useful interchangeable genetic parts, that could one day revolutionize genetic and synthetic biology. By the end of our project, we hope to double the number of yeast biobricks in the registry.
 "
"