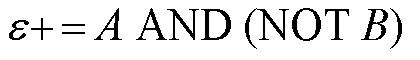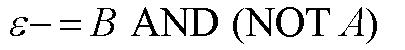Team:Minnesota/ProjectComparator
From 2008.igem.org
| The Minnesota iGEM '08 team consists of groups working on two different projects. One team is located primarily in Minneapolis and is designing a comparator for their project. The other team is based in St. Paul and is working on a time bomb. | |
|
Team Comparator | |
| Control systems are an integral component of almost all aspects of life. Whether it is in industrial, biological, or chemical applications, controllers provide a way to keep systems functioning properly. A vital part of any control system is the comparator. This component compares a set point value and a measured value, and determines which is larger. It then sends the appropriate signal to the controller, which reacts to bring the system back to the set point. In typical applications, the controller equipment is electronic. However, our team set out to create a comparator using only genetic components. This comparator could potentially be used as part of a new, solely biological control system that could be used to treat many diseases afflicting humans, for example diabetes. This comparator could compare the blood sugar of a patient to what it should be, and send this result to a control system that could compensate, for example by changing the levels of insulin.
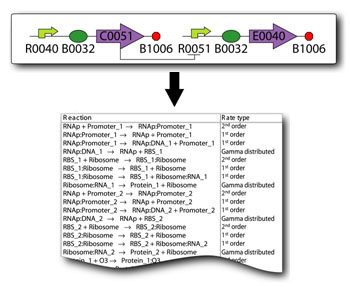
In order to undertake this task, a system involving six genes was designed. For our system, the two inputs (one representing the set point and one representing the measured value) are IPTG and aTc. These inputs will activate the transcription of the LacI and TetR proteins, and set in motion the rest of the system to produce the outputs. Depending on the amounts of the two inducer molecules added to the system, either green fluorescent protein (GFP) or red fluorescent protein(RFP) will be produced. The actual design of the system can be seen below. In order to complete this project, a total of six genes will have to be cloned into plasmids, and two new BioBrick parts will be produced. One will be a TetR and p22-MNT dual-repressed promoter, and the other will be a LacI and lambdaphage cI dual-repressed promoter. We will also build mathematical models and conduct computer simulations that will help with the designs. This project will pave the way for other parts of a true genetic PID controller to be produced, which could be an exciting scientific development in the near future. Computational Tools Team Comparator is also developing two tools named the "SynBioSS Designer" and "SynBioSS Wiki". SynBioSS stands for Synthetic Biology Software Suite. As with any engineering problem, modeling the behavior of a putative synthetic biological system is a key step in the design process. To date, many of these simulations of synthetic systems have used coarse-grained models and deterministic kinetic equations. Our team, however, strives to use stochastic models spanning multiple time scales. One major bottleneck is that the detailed molecular-level reaction networks required for such simulations are time consuming to construct. Moreover, while many of the promoters, operators, repressors, etc. used in synthetic biology have been well studied, the necessary quantitative information is scattered throughout the literature. The SynBioSS Designer facilitates the construction of detailed and accurate models, while the SynBioSS Wiki addresses the challenges of data collection and curation. | |
| Minnesota Home | Team Comparator Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Contents |
Computational Tools
SynBioSS Designer
SynBioSS Designer is an application for the automatic generation of sets of biomolecular reactions. This software allows a user to input the molecular parts involved in gene expression and regulation (e.g. promoters, transcription factors, ribosomes, etc.) The software then generates complete networks of reactions that represent transcription, translation, regulation, induction and degradation of those parts. To facilitate the creation of detailed kinetic models of synthetic gene networks composed of BioBricks, we have adapted SynBioSS Designer to automatically generate a kinetic model from a construct composed entirely of BioBricks. A NetCDF file is generated for simulations.
The web interface to the Designer, as well as an example of its use, can be found here[http://neptune.cems.umn.edu/designer/interface1.php].
SynBioSS Wiki
The inaccessibility of requisite kinetic data complicates the generation of detailed mechanistic models. We address this barrier by creating a web accessible database curated by users in a “Wiki” format. SynBioSS Wiki is a significant extension of the open-source Mediawiki software. The Wiki stores reaction kinetic data in a formatted and searchable scheme with references to the relevant literature. This framework allows for the input of reactions whose rates are described either by elementary first & second order rate equations or any arbitrarily complex rate equation defined using MathML (e.g. Hill type reactions). Reactions may be searched via participating molecules which may be proteins, DNA sequences, small molecules, etc. Once located, reactions of interest (along with their associated kinetic data) may be collected. The completed model may be exported in SBML format for additional editing or simulation.
It is also through the SynBioSS Wiki databases that SynBioSS Designer can access and proliferate kinetic information related to the simulation of BioBricks, thus extending the utility of the database for the benefit of the greater modeling community. To jumpstart the process, we have entered the known biomolecular interactions in the expression and regulation of well-studied operons, such as the lactose, the tetracycline and the arabinose operons.
The Wiki can be found here[http://neptune.cems.umn.edu/wiki].
Comparator Project
Control systems are an integral component of almost all aspects of life. Whether it is in industrial, biological, or chemical applications, controllers provide a way to keep systems functioning properly. A vital part of any control system is the comparator. This component compares a set point value and a measured value, and determines which is larger. It then sends the appropriate signal to the rest of the system. In typical applications, this system is electronic. However, our team set out to create a comparator using only genetic components. This comparator could potentially be used as part of a new, solely biological control system that could be used to treat many diseases afflicting humans, for example diabetes. This comparator could compare a diabetics blood sugar to what it should be, and send this result to a control system that could compensate.
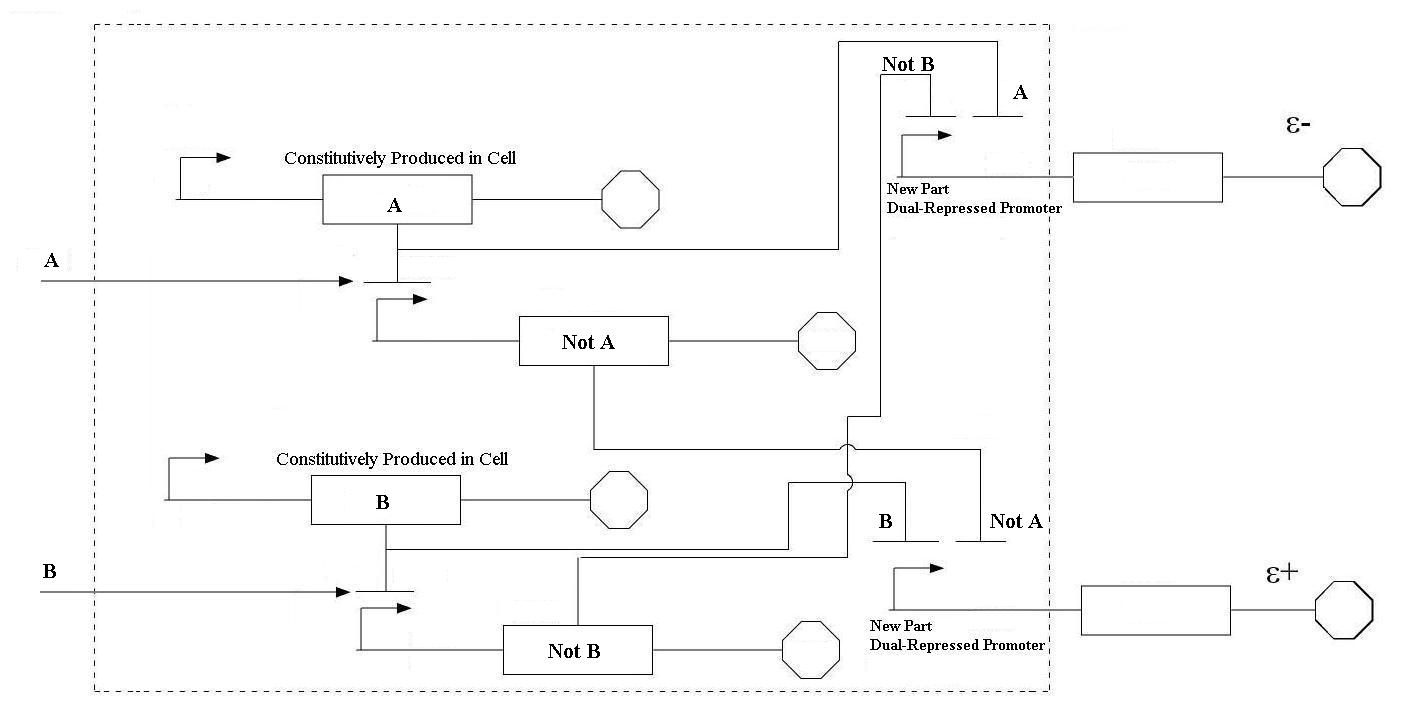
In order to undertake this task, a system involving six genes was designed. For our system, the two inputs (one representing the set point and one representing the measured value) are IPTG and aTc. These inputs will activate the transcription of the LacI and TetR proteins, and set in motion the rest of the system to produce the outputs. Depending on the amounts of the two inducer molecules added to the system, either green fluorescent protein(GFP) or red fluorescent protein(RFP) will be produced. The actual design of the system can be seen below. In order to complete this project, a total of six genes will have to be cloned into plasmids, and two new BioBrick parts will be produced. One will be a TetR and p22 mnt dual-repressed promoter, and the other will be a LacI and lambdaphage cI dual-repressed promoter. This project will pave the way for other parts of a true genetic PID controller to be produced, which could be an exciting scientific development in the near future.
Project Details
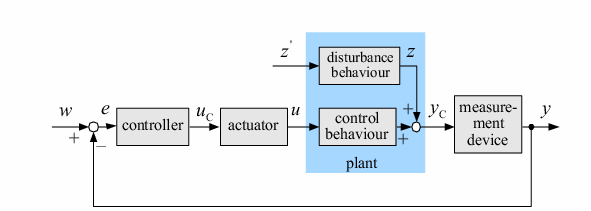
| In order to design a genetic comparator, the team first looked at an electronic comparator to try to emulate it. A typical comparator has two inputs: A is the set point value, which is input into the system, and B is the measured value, which is found by measuring the controlled variable and sending this result back to the comparator. Once the comparator recieves these two values, it compares them and produces a signal. Typical this signal is positive or negative. However, for a genetic controller, there is no such thing as a negative growth, so the genetic system was forced to have two outputs, instead of the one found on typical systems. These outputs were designated epsilon+ and epsilon- to distinguish whether the signal was positive or negative. A black-box comparator can be seen below. |
| The goal of our comparator design is to take two inputs and produce an output based on the level of each input. In order for this to be accomplished, we designed the comparator such that Epsilon = A - B, where A and B are the designated inputs. If A is larger, the epsilon+ response is observed, whereas if B is larger, the epsilon- response is observed. In the rare biological case where A and B are equal, no reponse will be seen, indicating that the system is at equilibrium -- the setpoint. In our system, the epsilon+ and epsilon- responses are differentiated by being linked to fluorescent proteins. This will allow us to readily test the effectiveness and accuracy of our comparator.
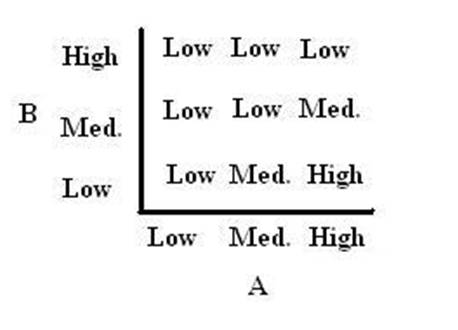
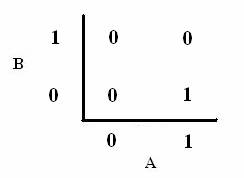
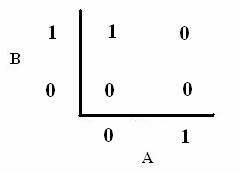
As shown below, the level of each output response can be qualitatively modeled. Partitioning the level of each input, A and B, into low, medium, and high quantities indicates the corresponding output response. Thus, only when the qualitative measurement of input B is less than that of input A will a large epsilon+ response be observed. The same applies in the case of the epsilon- response. This same idea can be translated into Boolean logic, as indicated below. Based on this logic, only when A is present and B is not, an epsilon+ response will occur. Similarly, only when B is present and A is not, the epsilon- response will be observed. |
|
From these Boolean tables, logic statements can be generated that give the response desired. These statements are shown below. |
|
Another way to state this logic, which makes it easier to design a system that generates this response, is shown below. |
| Even though these logic statements seem straightforward, the actual implementation of this logic in a biological system is actually fairly difficult. For the two inputs to the system, there are four possible states: neither A or B, more A than B, more B than A, and equal A and B. In order for the controller to be robust, it must respond correctly to each of these scenarios. This is accomplished by following the logic from the equations above. In order get the four components of the equations above (A, B, NOT A, NOT B), four genes were needed, one for each of these components. Furthermore, there must be two sets of two genes each that are linked together. NOT A and A must be linked, and NOT B and B must be as well. In order to get the NOT relationship, the gene products of A and B must repress the production of the NOT A and NOT B genes. |
| Diagram of dually repressed promoter |
| Diagram of dually repressed promoter |
Relevant Papers
|
Protocol Papers | |
| Bacteria Culture Protocols | |
| PCR Tutorial | |
| Subcloning | |
| Transformations | |
| Western Blotting 1 | |
| Western Blotting 2 | |
|
Basic BioBrick Design | |
| BioBrick Vector Design Paper | |
|
Combinatorial Promoters Papers | |
| Modular AND Gate | |
| cis Regulatory Input | |
| Combinatorial Promoters | |
| Directed Evolution of AraC | |
|
p22 mnt Papers | |
| DNA binding specificity of the Arc and Mnt repressors | |
| Bacteriophage P22 Mnt Repressor | |
| The Bacteriophage P22 Arc and Mnt Repressors | |
|
Light Sensor Papers | |
| Chevalier Light Sensor | |
| Young Light Sensor | |
Young Light Sensor (caged IPTG)
The ExperimentsResults
|
 "
"