Team:BCCS-Bristol/BSim-User-Guide
From 2008.igem.org
Contents |
BSim User Guide
BSimApp
BSimApp is a graphical interface that enables a user to view simulations. It can be used to help users familiarise themselves with the process of defining simulations, watch small-scale simulations or verify the initial conditions of large-scale simulations.
Typing
cd fileLocation (where fileLocation is the location the source code was extracted to, eg. c:\IGEM_2008) java bsim.BSimApp
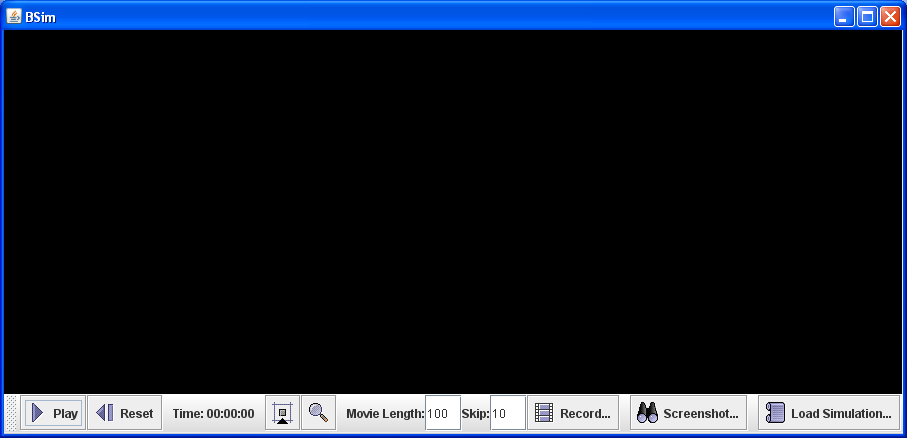
into the command prompt (Start -> Accessories -> Command Prompt) will run BSimApp and open up the graphical interface:
Menu
- Play / Pause - Plays or pauses the simulation
- Reset - Stops the current simulation and reverts to the initial conditions defined in the parameter file
- Video Length - Length of video to be recorded (frames)
- Frame Skip - Number of time steps to be skipped per frame
- Record Video - Records a video with properties defined in Video Length and Frame Skip
- Screenshot - captures the current state of the simulation in a .png picture file
- Load Simulation - The large black area is where the initial conditions of a simulation will appear once a parameter file has been loaded. Several example parameter files are supplied below and can be loaded by pressing the ‘Load Parameters’ button. The simulations defined by these files are summarised below:
- [[BSimExp-Density 300.txt Basic Chemotaxis]] - A simulation of basic chemotaxis from left to right with the 300 bacteria and 3 particles. the top and bottom boundaries of the simulation space are wrapping and the path of the particles are followed by tracers.
- [[Image:BSimExp-Adhesion 6.txt Adhesion]] - 6 chemotactic bacteria are adhered to the particle. There are no boundaries to the simulation space and the bacteria can be viewed by zooming in on the particle. The particle's position is also followed by a tracer.
- [[BSimExp-Recruit_Density_100_10_min.txt Recruitment]] - Shows 100 recruitment bacteria and one particle. The bacteria change colour depending on their state:
- Green - Random movement, the bacteria move randomly with no bias.
- Blue - Contact, the bacteria release a recuitment signal
- Yellow -
- Red -
The process of using parameter files to define your own simulation is explained here.
BSimBatch
Due to the high levels of randomness in the run and tumble motion of bacteria it is likely that the user will want to run the same simulation many times and use statistical analysis to investigate emergent behaviour. BSimBatch enables the user to run batches of simulations, saving time series of the mean bacteria position and particle positions for each simulation.
BSimBatch is run using the command:
cd c:\IGEM_2008 java bsim.BSimBatch ParameterFileLocation
Where ParameterFileLocation is the location of the parameter file defining simulation to be run, eg:
java bsim.BSimBatch ./Parameter_Files/Parameter_File_1.txt
Time series relating to the positions of the particles and mean position of bacteria will be outputted to the location defined in the parameter file.
Problem Solving
BSim is written in the [http://en.wikipedia.org/wiki/Java_programming_language Java programming language] so any computer using BSim must have a [http://en.wikipedia.org/wiki/Java_Runtime_Environment Java environment] before any simulations can be run; [http://www.jibble.org/settingupjava.php This page] has a nice guide on setting up a Java environment.
 "
"