From 2008.igem.org
Recipes
- Tris-Cl, 1M
- Dissolve 121g Tris base in 800ml H2O
- Adjust to desired pH with concentrated HCl
- Mix and add H2O to 1 liter
- (Approximately, 20ml HCl for pH 7.4 and 42ml for pH 8.0)
- EDTA, 0.5M (pH 8.0)
- Dissolve 186.1g Na2 EDTA-2H2O in 700ml H20
- Adjust pH to 8.0 with 10M NaOH(~50ml)
- Add H2O to 1 liter
- Breaking buffer - 100ml
- 2ml Triton X-100
- 1ml Sodium dodecyl sulfate (SDS)
- 0.5844g NaCl (100mM)
- 1ml 1M Tris-Cl pH 8.0 (10mM)
- 200uL 0.5M EDTA (1mM)
Primers
PGK promoter
5'TTTT GAATTC AAAGATGCCGATTTGGGCGC 5'TTTT GAGCTC GTTTTATATTTGTTGTAAAA
PGK terminator
5'TTAT GGGCCC GAAATAAATTGAATTGAATT 5'TTTTG AAGCTT CAGCTTTAACGAACGCAGA
Ste2
5'GCCC TCTAGA ATGTCTGATGCGGCTCCTTC 5'TTAT GGGCCC TCATAAATTATTATTATCTT
Fus1 upstream
5'GTGG GAATTC TAATAATCAGAACTCCAACA 5'GGCG TCTAGA TTTGATTTTCAGAAACTTGA
Fus1 downstream:
5'GCGA GGTACC TGAAAATAATATTGACGTTC 5'TTAT GCGGCCGC TATTCACCAGACCCGCTCCT
22nd July
- Yeast obtained from Dr. Zhao
24th July
- Prepared liquid culture for DNA extraction
- Made 1M Tris. Cl pH 8.0
- Made 4M ammonium acetate
22nd August
- Attempted DNA extraction of W303a genomic DNA
- Protocol from Wiley's Current Protocols in Molecular Biology
- Result: Failed to finish protocol
- Obtained more yeast from Dr. Zhao
25th August
- Prepared overnight culture for DNA extraction (3:27pm)
26th August
- Attempted DNA extraction
- Prepped overnight culture
27th August
- Performed PCR: PGK Terminator
| Buffer G
| 12.5uL
| x4
| 50uL
|
| Forward Primer
| 0.5uL
| x4
| 2uL
|
| Reverse Primer
| 0.5uL
| x4
| 2uL
|
| H2O
| 10.8uL
| x4
| 43.2uL
|
| Taq
| 0.2uL
| x4
| 0.8uL
|
| template
| 0.5ul
|
| Negative control
| 3 H2O
|
- PCR program:
- 4 min 94 degrees
- 25-30x 30s 94 degrees
- 30s Tm primers
- 1 min/KB 72 degrees
- 7 min 72 degrees
- For the gel: 5uL loading dye gel is in cold room
- Prepped 3 overnight cultures
28th August
- Extracted DNA from 4 cultures
- Ran gel of PCR products (1.5% agarose, 200V)
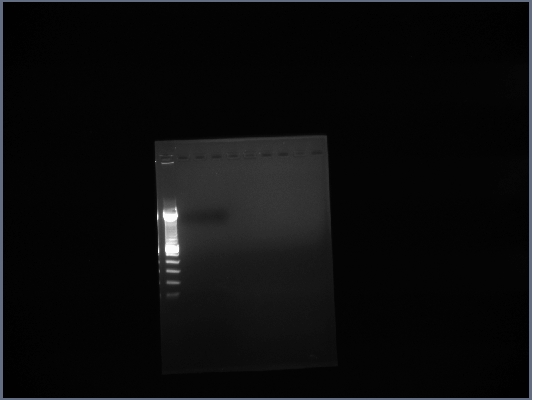
2nd September
| 5 PRIME Mastermix
| 10uL
| x3
| 30uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template
| 10uL
| x3
| 30uL
|
| H2O
| 10.8uL
| x4
| 43.2uL
|
| Negative control
| 25uL H2O
|
3rd September
- Ran gel
- Ladder lane 7
- Sample 7 spilled
- 1% agarose
- 120V
- 50 minutes
- Poor results--no bands present
8th September
| 5PRIME Mastermix
| 10uL
| x3
| 30uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template
| 14uL
| x3
| 42uL
|
- Prepped 4 overnight cultures
9th September
- Signs of life in 3 of the cultures
- Ran gel on PCR from 8th September--no bands present
- 150V, 50 minutes
- No sign of DNA
- Ladder from Courtney
10th September
- Split culture
- Ran gel from 8th September again--no bands present
- 150V, 50 minutes
- 0.75% gel
- Ladder from Courtney
11th September
| 5 PRIME Mastermix
| 10uL
| x3
| 30uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template
| 14uL
| x3
| 42uL
|
12th September
- Isolated genomic DNA from 8 cultures of W303a yeast cells
- Protocol from Wiley's Current Protocols in Molecular Biology
- Labeled templates 1,2,3,4 and 1a,2a,3a,4a
- Ran gel of PCR from 11th September
15th September
- PCR PGK Promotor
- Finnzymes Phusion High Fidelity DNA Polymerase
| 5x Phusion HF Buffer
| 10uL
| x3
| 30uL
|
| 10mM dNTPs
| 1uL
| x3
| 3uL
|
| Primer A(Forward)
| 1uL
| x3
| 3uL
|
| Primer B(Reverse)
| 1uL
| x3
| 3uL
|
| Template 1
| 10uL
| x3
| 30uL
|
| Phusions DNA polymerase
| 0.5uL
| x3
| 1.5uL
|
| H2O
| 26.5uL
| x3
| 79.5uL
|
18th September
- Ran reaction mentioned on 15th September
- Extracted DNA from gel from 8th September (PGK Terminator)
| Tube
| Gel(g)
|
| 1
| 0.332
|
| 2
| 0.278
|
| 3
| 0.307
|
| 4
| 0.349
|
| 5
| 0.385
|
19th September
- Gel of PGK Promotor from 15th September has no DNA present
23rd September
- PCR: Fus1 Downstream
- EPICENTRE Bioetechnologies - MasterAmp(TM) Taq DNA Polymerase
| MasterAmp Taq 10x PCR Buffer
| 5uL
|
| 1mM dNTPs
| 1uL
|
| Primer 1
| 0.5uL
|
| Primer 2
| 0.5uL
|
| 25mM MgCl2
| 2uL
|
| Taq DNA Polymerase
| 0.25uL
|
| Template 2
| 20uL
|
| Water
| 20.75uL
|
- PCR Settings:
- 4mins, 94 degree celcius
- 30s, 94 degree celcius
- 30s, 5 degrees below primer melting temperature
- 1 min, 72 degree celcius -- to step 2 -- 30x
- 7 min, 72 degree celcius
24th September
- Ran gel of Fus1 Downstream from 23rd September
- Result: No DNA present on gel
25th September
| Mastermix
| 8.25uL
| x3
| 24.75uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template 3
| 20uL
| x3
| 60uL
|
| Water
| 20.75uL
| x3
| 62.25uL
|
- same protocol as 23rd September
- The master mix contains the buffer, dNTPs, MgCl, and Taq
- Template 3 (3 reactions run)
30th September
| Mastermix
| 8.25uL
| x3
| 24.75uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template 3
| 20uL
| x3
| 60uL
|
| Water
| 20.75uL
| x3
| 62.25uL
|
- same protocol as 23rd September
- Template 4 and 1a (3 reactions each)
- Gel: 1% agarose, 150V, 35 minutes -> poor results
- lanes 2,3 -> faint smear

1st October
| Mastermix
| 8.25uL
| x3
| 24.75uL
|
| Forward Primer
| 0.5uL
| x3
| 1.5uL
|
| Reverse Primer
| 0.5uL
| x3
| 1.5uL
|
| Template 3
| 20uL
| x3
| 60uL
|
| Water
| 20.75uL
| x3
| 62.25uL
|
- same protocol as 23rd September
- Templates 2a, 3a, 4a used (3 reactions each)
3rd October
- Ran Ste2 gel from 1st October
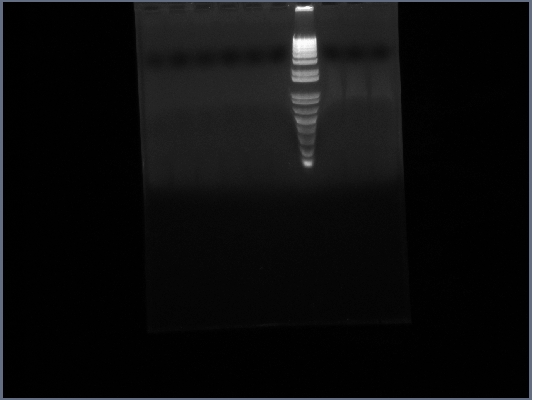
8th October
| PCR Buffer
| 5uL
|
| 10mM dNTPs
| 1uL
|
| Forward Primer
| 1uL
|
| Reverse Primer
| 1uL
|
| MgCl2
| 5uL
|
| Taq DNA Polymerase
| 0.25uL
|
| Template
| 25uL
|
| Water
| 11.75uL
|
- same protocol as 23rd September
- Use DNA extracted from gel on 18th September (5 reactions)
- Also extracted DNA from gel from 30th September (Fus1 Upstream)
9th October
- PCR: Ste2
- Protocol is the same as 23rd September
- Template used is product from 1st October (9 reactions total)
- PCR: Fus1 Upstream
- Protocal matches 23rd September
- Template used was DNA extracted from the gel from the 8th October, which came from the PCR run on 30th September (5 reactions total)
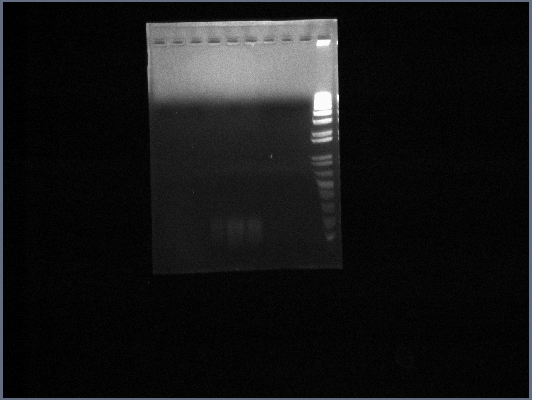
- Ran PGK terminator gel from yesterday
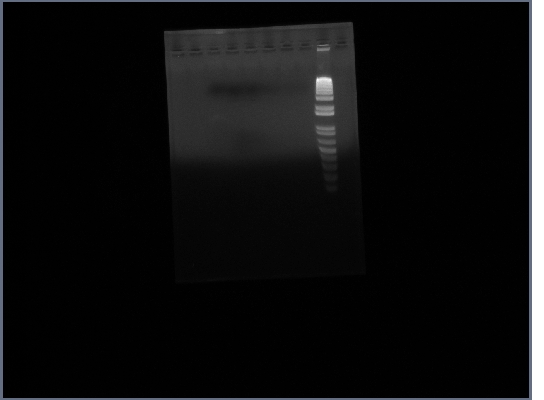
12th October
- PCR: Fus1 (3 reactions each)
- Template is products from 9th October
13th October
- Ran gel of PCR with Fus1
- Used 25uL or product, 5uL of loading dye

14th October
| MasterAmp Taq 10x PCR Buffer
| 5uL
|
| 1mM dNTPs
| 1uL
|
| Primer 1
| 0.5uL
|
| Primer 2
| 0.5uL
|
| 25mM MgCl2
| 5uL
|
| Taq DNA Polymerase
| 0.25uL
|
| Template
| 25uL
|
| Water
| 11.75uL
|
- Template is reaction from 12th October
15th October
| 5 PRIME MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 25uL
|
| Water
| 4uL
|
- Ran gel of Ste2 from 10/14
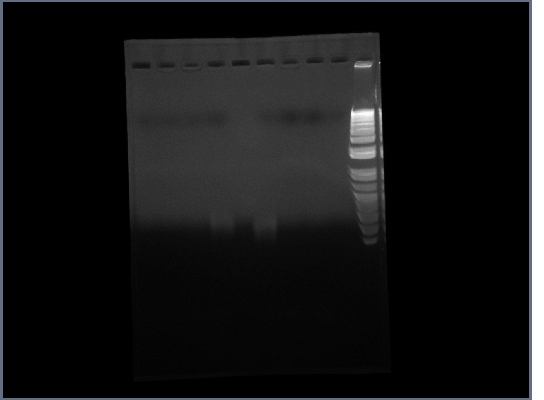
- Ran gel of Fus1 upstream from 10/14 (no ladder, oops)
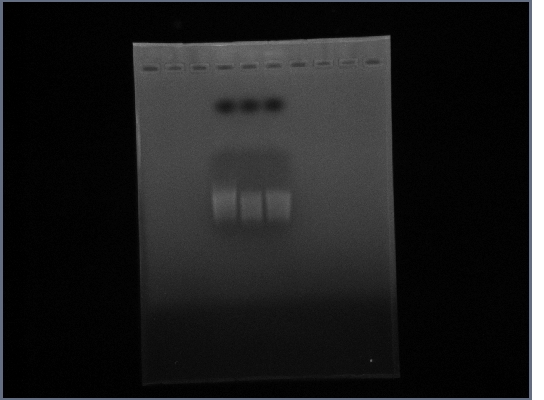
16th October
- PCR: Fus1 Downstream, PGK Promoter
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
- Tube 1: Template 4 - Fus1
- Tube 2: Template 4a - Fus1
- Tube 3: Template 4 - PGK Promoter
- Tube 4: Template 4a - PGK Promoter
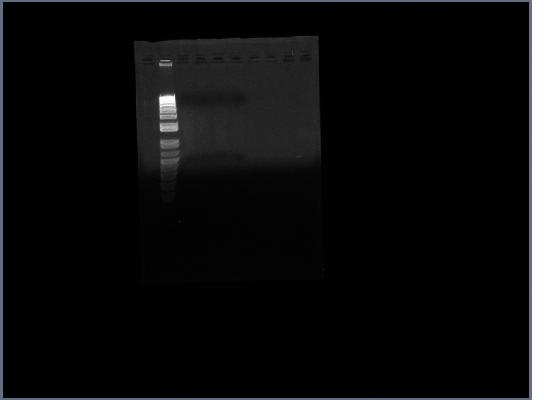
- ran Gel of PGK terminator from 10/15
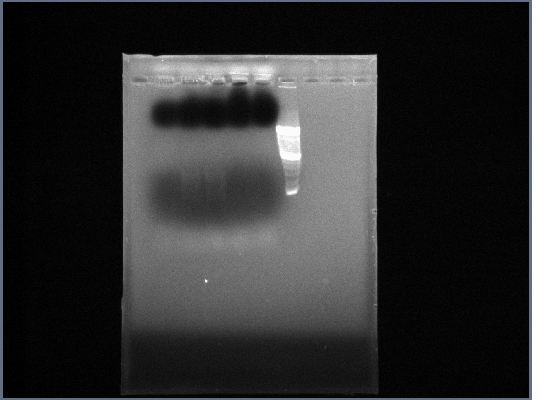
- Extracted Ste2 from 10/15 and re-amplified:
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
17th October
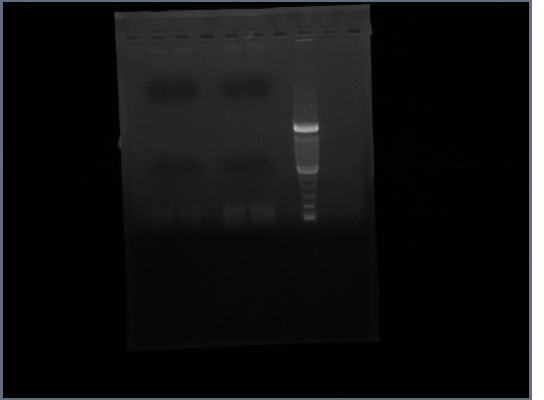
- Ran gel of Ste2 from 10/16
- Tube 1 - lane 2,3
- Tube 2 - lane 5,6
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
- template is products from 10/14
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
- Template is products from 10/15
- Extracted Ste2(Gel from today)
18th October
Ran gel of Fus1 upstream and PGK terminator from yesterday
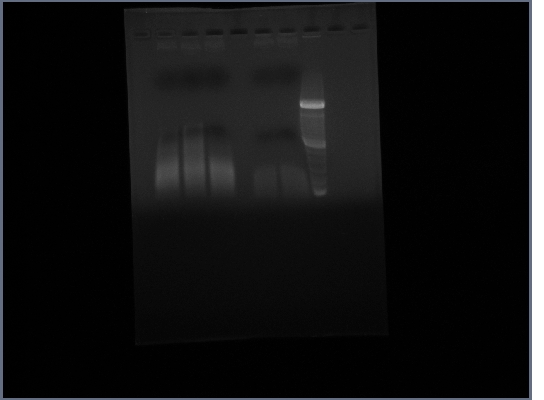
20th October
- Extracting chromosomal DNA from yeast cells
- W303A - Yellow
- YPD DL
- YHP1 YPD HD - One is orange (YHP1)(Cap was removed in incubator), Other is Yellow
- Orange - YHP1 YPD HD
- Green - YHP2 YPD HD
- Pink - W303A YPA DL
- Yellow - WD303A YPD DL
21st October
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
- 2 tubes
- Block B of black machine
- Annealing temperature: 31 degrees celcius
- Ran gel on Ste2 from today
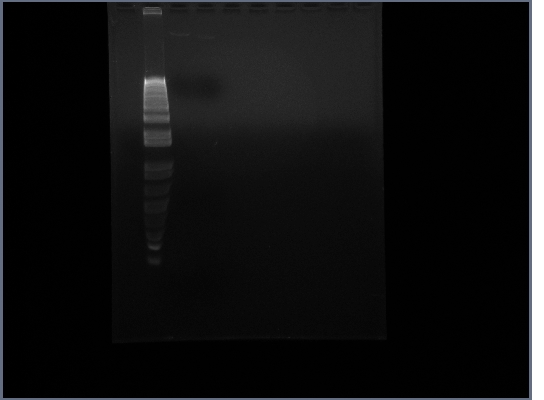
22nd October
- New primers for biobricks are here
- Brought to standard concentration, 30uM
- Added 33.3uL of water per nmol primer
- PCR: Fus1 and PGK Terminator
| MasterMix
| 20uL
|
| Primer Fwd
| 0.75uL
|
| Primer Rev
| 0.75uL
|
| Template
| 25uL
|
| Water
| 3.5uL
|
- Forward and Reverse primers are new biobrick primers that arrived today
- Template is PCR product from 17th October
- A,B,C -> Fus1 Upstream -> 1,2,3
- 1,2 -> PGK Terminator -> 4,5
- Annealing temperature: 38 degrees celcius
- In freezer in yellow case
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 25uL
|
| Water
| 4uL
|
- Forward primer, Reverse Primer are new primers that arrived today
- Template is PCR product from 21st October
- Two tubes in block B
- The 3 gels in the cold room do not have EtBr
23rd October
- Ran gel of Fus1 Upstream, PGK Terminator, Ste2 from yesterday
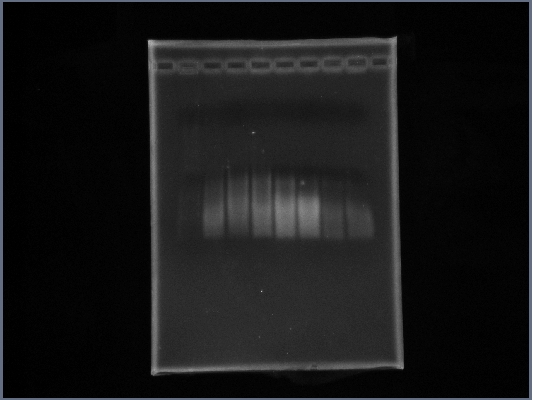
- PCR: Fus1 Downstream, PGK Promoter
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
- 4 reactions each of Fus1 Upstream(1,2,3,4) and PGK Promoter(A,B,C,D)
- Gel shows no bands.
- PCR: Fus1 Downstream, Fus1 Upstream, PGK Promoter, PGK Terminator, Ste2
- Template genomic DNA from 20th October (x4 different reactions)
- Used all Template
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 20uL
|
| Water
| 9uL
|
24th October
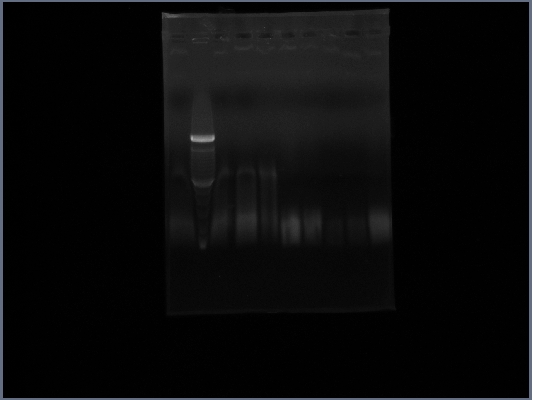
- Ran gel of Fus1 upstream(lanes 3,4,5), PGK terminator(lanes6,7), and Ste2(lanes 8,9) from 10/22
- ladder is lane 2; 1 and 10 are nothing
- PCR of Fus1 downstream, Fus1 upstream, PGK promoter, PGK terminator, Ste2
| MasterMix
| 20uL
|
| Primer Fwd
| 0.5uL
|
| Primer Rev
| 0.5uL
|
| Template
| 29uL
|
- The templates were the rest of the genomic DNA extracts from 9/12
- Three reactions of each gene were run
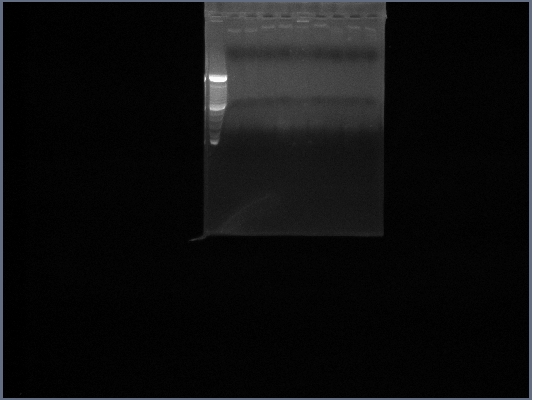
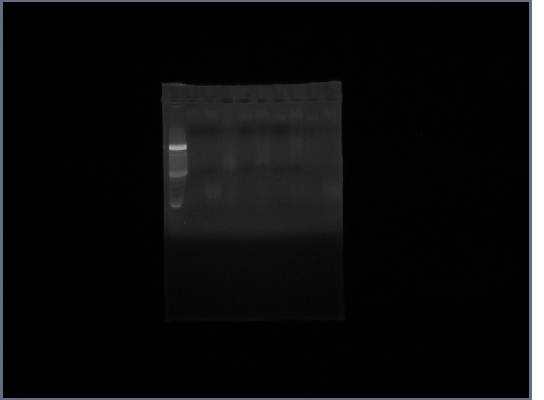
- (on left)Fus1 downstream lanes 2,3,4; Fus1 upstream lanes 5,6,7; PGK promoter lanes 8,9,10;
- (on right)PGK terminator lanes2,3,10; Ste2 lanes 4,5,6; Fus1 downstream lanes 7,8,9;
25th October
- Extracted DNA from the gel from 10/24
- FUS1 upstream from the higher bands of lanes 3 and 4
- PGK terminator from the lower bands of lanes 6 and 7
- DNA ligation
| DNA
| 5uL
|
| buffer
| 5uL
|
| Re1 (Pst1)
| 1uL
|
| Re2 (EcoR1)
| 1uL
|
| Water
| 37.5uL
|
26th October
- Incubate for 20mins at 80 degrees celcius
| Ligation Buffer
| 4uL
|
| DNA Ligase
| 1uL
|
| DNA
| 3uL
|
| Plasmid
| 9uL
|
| Water
| 3uL
|
- Let sit for 5 min.
- 5uL of above mixture to competent cells
- Heat shock 30s (42 degrees)
- Add SOC Media (200uL)
- Incubate 60 min.(37 degrees)
- Plate 200uL
- Incubate 37 degrees celcius overnight
27th October
The transformation failed; try again with the same protocol:
- Using extracts 3 and 7 (+Ligation buffers, Ligase, Plasmid, and Water)

 "
"
















