Team:University of Washington/SeToB
From 2008.igem.org

| Home | The Team | The Project | Modeling | Notebook | Protocols | Parts Submitted to the Registry | Measurement Kit | SeToB | Safety |
|---|
Summary
To describe the software project in a few words, I will say the main objective is to initiate a creation of registry of standard parts that is organized, accessible, programmable end point, and ease of use. Another objective is that given such a data store, we can build very interesting applications on top of it (one such example is described).
Motivation
Say, if one has a general circuit in mind, all one needs is then just parts satisfying his specifications on the circuit. Specifically, say I want to build a circuit involving two pairs of RNA, a arabinose induced promoter, and all other parts that will complete the picture (terminators, RBS, and so forth). Being a computer engineer (having very little biology knowledge), I go to the parts website and then search for all the parts one by one, opening tons of firefox (or opera, or chrome, or IE, or lynx for the sake of completion) tabs... All this is so complicated. Why can't it be much simpler, such as say few clicks aways!
Details
- We used PoBoL (http://pobol.org) as a data store for our project. It turns out that storing BioBrick parts in ontology format (OWL --> RDF --> N3 turtle) is very useful. The main advantages:
- Whatever are our advantages of using OWL, that is we can get a very basic reasoning unit.
- The files are serializeable (easily transmitted without any strings attached) over the web, and can be hosted at many end points (at the host's convenience), leading to a decentralized storage.
- Although PoBoL is in its minimal format right now, we needed a way to add sequence feature information to actually query to see if a promoter is repressed by pTet, what protein does a gene codes for by going to http://sequenceontology.org.
- The rest is just basic UI in Flash, and server side in Python.
Okay, so what did we do?
- We implemented a registry parser tool that is currently hosted at http://students.washington.edu/paramsan/tmp/setob/html/index.html.
- We converted some files into PoBoL format and stored them at http://students.washington.edu/paramsan/tmp/setob/bb_files/.
- We implemented a basic registry searcher end-point in Flash that pretty much causes the server to run SPARQL queries on the stored PoBoL files here http://students.washington.edu/paramsan/tmp/setob/flash/.
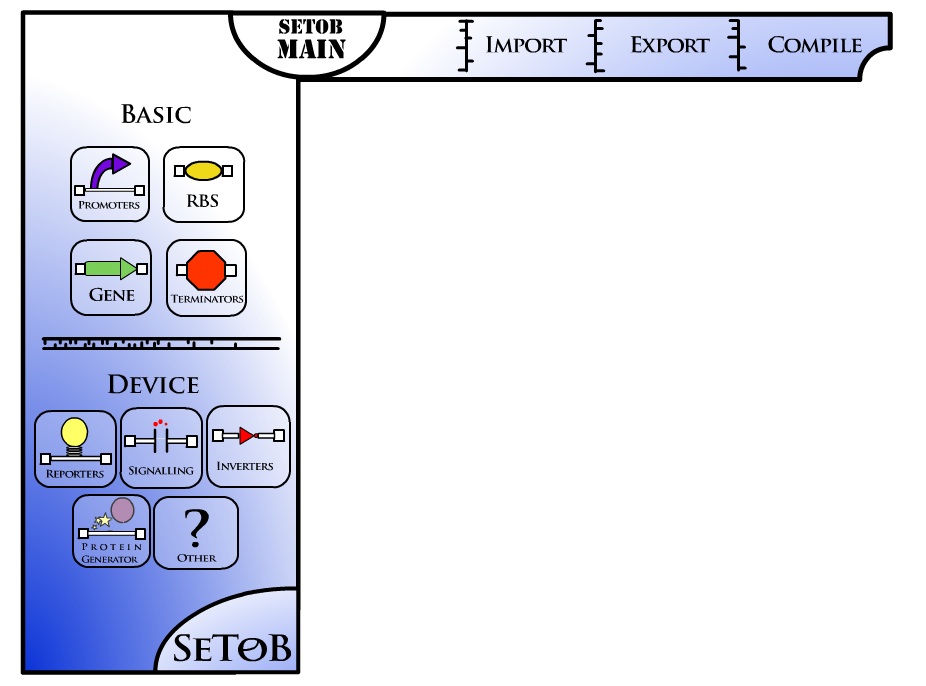
Here is what the Flash UI prototype looks like (check for updates on http://setob.blogspot.com)
| Figure 1. Flash UI Prototype, the parts can be dragged onto screen and "prospective" parts should show up after the part is in the white area. |
Blog
The overwhelming discussion (sarcastically) for the software tool is hosted at http://setob.blogspot.com.
Code Repository
The code is licensed under MIT License (the openest of all at Google code in my opinion) at http://setob.googlecode.com.
SeToB?
The project was initially designed to be a schematic to a BioBrick parts convertor (given a bio-circuit, find the BioBrick parts that will fit the criterion. However, it was later changed to just find parts given another part.
Future
Assuming we have the basic software working, we can then go on for many wild adventures, such as,
- Implement the schematic input idea (convert schematic to BioBricks)
- Implement some first order logic ideas, that is converting a circuit requiring AND, NOT, OR (and their derivatives) operations.
- Given a composite part that the software help create, list the protocols that can be used to physically build the part.
Team Members
- Students
- [http://students.washington.edu/paramsan/ Paramjit Singh Sandhu]
- Tyler Casey
 "
"