Team:Hawaii/Large-Scale Preparation of Plasmid from E. coli
From 2008.igem.org
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
Large-Scale Preparation of Plasmid DNA
A protocol for the preparation of plasmid DNA from large cultures. Adapted from a protocol for a 500mL prep. Yields milligram quantities of reasonably clean crude DNA. To obtain highly purified plasmid DNA can be obtained by using CsCl/ethidium bromide equilibrium centrifugation (protocol found in Maniatis).
Also included are two assays for verification and quantification of plasmid DNA: UV wpectrometric quantification and restriction digest followed by gel electrophoresis.
Methods
Large-Scale Plasmid Prep Procedure Materials
- LB medium containing selective agent
- Plasmid bearing E. coli strain
- GTE solution (50mM Glucose/10mM EDTA/25mM Tris-HCl, pH 8.0)
- NaOH/SDS solution (0.2M NaOH/1.0%SDS)
- 10mg/mL Rnase in Tris-HCl, pH 7.5
- Isopropanol
- 70% (v/v) ethanol
- centrifuge
- 2L baffled flask
- 50mL centrifuge tubes
- Water Bath at 55°C
- Innoculate 5mL LB containing selective agent with a colony of plasmid bearing E. coli. Grow at 37C with vigorous shaking over night.
- Inoculate 500mL LB containing selective agent with ~1mL of over night culture. Grow at 37C with vigorous shaking until OD600~4.0 is reached (saturation).
- use baffled 2L flask
- for this prep, we used a 300mL culture.
- Centrifuge 10 minutes at maximum 894 rcf (maximum rcf for our centrifuge), 4°C. Use 50mL aliquots.
- The protocol recommends using 6,000 x g.
- The 300mL prep is divided into 6 50mL tubes.
- Combine 3 tubes by resuspending in 4mL of GTE solution. Incubate 10 minutes at room temperature.
- For one variation (latter called Prep 1), add 50ug/mL Rnase solution (20uL).
- Add 10mL NaOH/SDS solution, mix (gently) by inverting 4 times. Incubate on ice for 10 minutes.
- Solution should become homogeneous and clear. This prep was not clear, but proceeded with protocol.
- Add 7.5mL potassium acetate, mix (gently)by inverting 4 times. Incubate on ice for 10 minutes.
- A white precipitate forms.
- Centrifuge for 15 minutes at 894 rcf, 4°C.
- Recommended to spin at 20,000 x g for 10 minutes.
- Centrifuge until a good pellet forms. Some material will be floating, remove as much as possible with pipette tip.
- Decant supernatant to a new tube.
- Do this step with a pipette and avoid the white precipitate.
- For one variation (latter called Prep 2), add 50ug/mL Rnase solution (20uL).
- Add 0.6 volume of isopropanol. Mix by inversion, let stand 5-10 minutes at room temperature.
- For a 21.5mL prep (total volume up to this point) add 12.9mL isopropanol.
- Centrifuge 15 minutes at 894 rcf at room temperature. Discard supernatant.
- Recommended to spin at 15,000 x g.
- Centrifuge until really good pellet forms. Avoid the pellet!
- Wash pellet with 2mL 70% ethanol.
- Centrifuge for 5 minutes at 894 rcf at room temperature. Aspirate ethanol.
- Recommended to spin at 15,000 x g briefly.
- Centrifuge until really good pellet forms.
- Dry pellet in the hood.
- Recommended to dry the pellet under vacuum.
- Resuspend in 100uL TE, lightly vortex.
- Recommended to store indefinitely at 4°C (but does not specify if buffer is needed).
- Heat at 65°C for 30 minutes.
- The product was cloudy so taking extra purification step.
- Centrifuge 10 minutes at 894 rcf, room temperature.
- Aspirate clear liquid, avoiding pellet.
- Wash pellet with 100uL TE, centrifuge, aspirate and combine with product.
- Check the concentration of the plasmid using a spectrophotometer (need protcol).
- Verify presence of plasmid DNA by first using a restriction digest, followed by gel electrophoresis.
UV Spectroscopy for the Quantification of Plasmid DNA
- used to asses purity and concentration of nucleic acids
- A260 measurements are quantitative for relatively pure nucleic acid preps in microgram quantities
- Cannot be used to discriminate between RNA and DNA
- Ratio of A260/A280 indicates purity, as protein absorbs at 280nm.
- A325 indicates particulates in solution or dirty cuvette
- A230 for contaminants containing peptide bonds or aromatic moieties such as protein and phenol
Materials
- 1x TE buffer
- nanopure water
- plasmid prep (for concentrated preps, need several dilutions)
- pipetter (for 2uL quantity)
- tips
- nanodrop spectrophotometer
- chem-wipes
Procedure
- Turn on computer, select spec icon, choose nucleic acids setting
- Pull up lever of spec (DON'T PULL with WIRE!), wash top and bottom with small amount of water.
- Blank with 2uL TE, click blank
- Load 2uL sample, click measure
- If results significant, can print. If not, repeat steps with a dilution of sample.
Procedure for a 10uL Digestion/Gel Electrophoresis Materials
- EcoRI
- EcoRI buffer
- BSA
- nanopure water
- pRL1383a (from prep 1 and prep 2)
- heat block at 37°C (later 65°C)
- 1.5 mL centrifuge tubes
- agarose gel
- gel apparatus
- Ethidium Bromide
- 6X loading buffer
- Ladder
- Turn on heat block.
- To the centrifuge tube, add 2uL nanopure water, 1uL EcoRI buffer, 1uL BSA, 5uL pRL1383a plasmid prep.
- Microcentrifuge until all contents are in solution at bottom of tube.
- Add 1uL EcoRI.
- Place on heat block for 1 hour.
- While waiting, prepare the gel.
- Microwave the gel in bottle in 30 second intervals, mixing in between until all of gel is melted.
- add Ethidium Bromide to gel.
- There should be some already in the gel, adjust as needed.
- Put in well clip, pour gel after it is sufficiently cooled (does not hurt when you touch it).
- After 1 hour, increase heat on block to 65°C and incubate digestion for 20 minutes.
- Heat inactivation of EcoRI.
- Prepare samples for gel electrophoresis:
- Plan out wells, in addition to digestion, add circular plasmid as a control.
- Mix sample + loading buffer in 5:1 ratio.
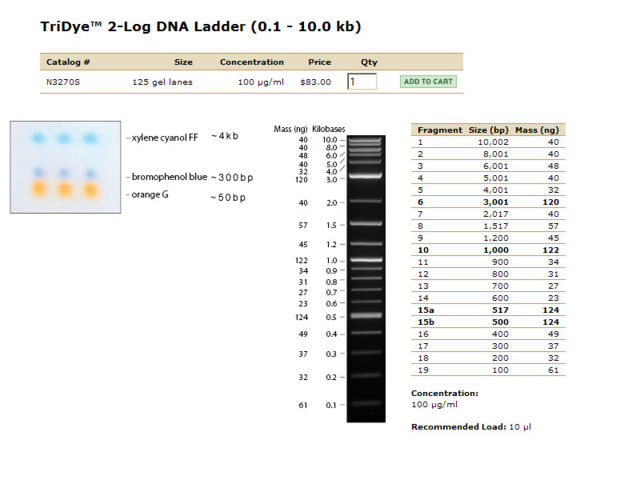
- Load Ladder ([http://www.neb.com/nebecomm/products/productN3270.asp NEB 2-log Tri-dye Ladder]) and samples.
- Run gel at 95V for 45 minutes or until end of ladder reaches halfway down gel.
- Take some pictures.
Results
UV Spectroscopy for the Quantification of Plasmid DNA
| (Prep#)Dilution | A260 | A280 | A260/A280 | Amount (ng/uL) | A260/A230 |
|---|---|---|---|---|---|
| (Prep1)1:10 | 52.514 | 26.854 | 1.96 | 26257.0 | 2.38 |
| (Prep1)1:100 | 11.048 | 5.745 | 1.92 | 55240.0 | 2.42 |
| (Prep1)1:1000 | 1.25 | 0.619 | 2.02 | 62500.0 | 2.71 |
| (Prep2)1:10 | 62.512 | 32.703 | 1.91 | 31250.0 | 2.34 |
| (Prep2)1:100 | 4.58 | 2.434 | 1.96 | 23790.0 | 2.36 |
| (Prep2)1:1000 | 0.597 | 0.288 | 2.07 | 29900.0 | 3.62 |
Electrophoresis Results
| Lane | Contents | Description | Amount (ng/10uL) | Size | Amount RNA (brightness of smear in low kb region) |
|---|---|---|---|---|---|
| Lane 1 | The ladder | Faint, but sufficiently separated | |||
| Lane 2 | Prep 1, no dilution | Most of the DNA appears to have remained in the well, a smear indicates some has run to about 10kb. | |||
| Lane 3 | Prep 2, no dilution | Most of the DNA appears to have remained in the well, a faint smear indicates some has run to about 10kb. | |||
| Lane 4 | Prep 1, 1:10 dilution | DNA appears to have remained in the well. | |||
| Lane 5 | Prep 2, 1:10 dilution | DNA appears to have remained in the well. | |||
| Lane 6 | Prep 1, 1:100 dilution. | No nucleic acids stained. | |||
| Lane 7 | Prep 2, 1:100 dilution | No nucleic acids stained. |
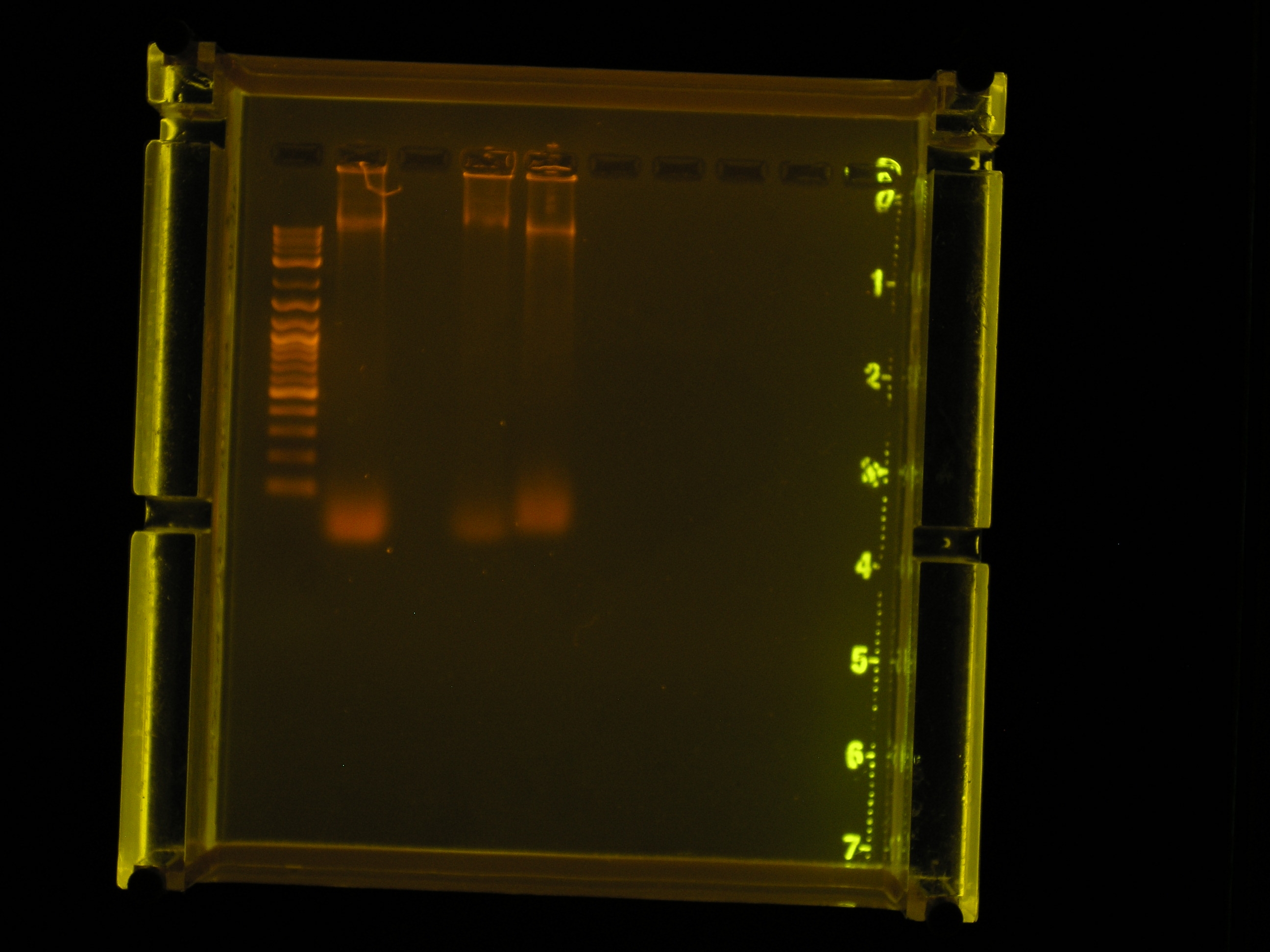
| Lane | Contents | Description | Amount (ng/10uL) | Size | Amount RNA (brightness of smear in low kb region) |
|---|---|---|---|---|---|
| Lane 1 | The ladder | Sufficiently separated | |||
| Lane 2 | Prep 1 Circular DNA (Control) | One bright band in high bp region, one bright band near 10kb region, large smear in low kb region. | 34ng | ~10kb | 124ng |
| Lane 3 | Prep 1, Linear DNA | No nucleic acids stained. | |||
| Lane 4 | Prep 2 Circular DNA (Control) | One semi-bright band in high bp region, one bright band near 10kb region, small smear in low kb region. | 32ng | ~10kb | 61ng |
| Lane 5 | Prep 2, Linear DNA | One bright band in high bp region, one bright band near 10kb region, small smear in low kb region. | 49ng | ~10kb | 49ng |
Discussion
- A large scale plasmid prep can be performed with this procedure.
- From the gel electrophoresis results, is not apparent that the step in which the Rnase is added is important because a smear in the low kb region is found in both preps indicating a possible RNA contamination. We should add as the manual says after the GTE addition so that the experiment can follow this manual as closely as possible.
- To improve the RNA digestion, we will adjust the protocol by incubating the RNase for 30 minutes at 55°C.
- From the gel electrophoresis results, we can conclude that it is necessary to linearize the plasmid DNA before a gel is run. It is also apparent that levels of Ethidium Bromide should be added so that sufficient staining is achieved.
- This protocol needs to be updated with the UV Spec quantification experiment and the concentrations calculated (they are locked in the lab right now!).
References
- "Large-Scale Preparation of Plasmid DNA,"Short Protocols in Molecular Biology, published by John Wiley & Sons, Fifth Edition, Volume 1, pages (1-25)-(1-26).
- "Detection of Nucleic Acids using Absorption Spectroscopy,"Short Protocols in Molecular Biology, published by John Wiley & Sons, Fifth Edition, Volume 2, pages (A3-16)-(A3-17).
Insanity is doing the same thing over and over again and expecting different results. - Albert Einstein
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"