Team:Hawaii/PCR Amplification of pRL1383a
From 2008.igem.org
Revision as of 23:53, 4 July 2008 by MargaretRuzicka (Talk | contribs)
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
PCR Amplification of pRL1383a
- Several regions of pRL1383a will be amplified with BioBrick based primers. These components will be used later in the construction of a pRL1383a BioBrick based vector. These parts include the aadA region of the omega interposon, the origin of vegetative replication (oriV), the mobilization proteins, and the replication proteins.
Methods
Materials Per Reaction Added in order:
- 3.5uL nanopure water
- 1uL 10uM forward/reverse primers
- 1uL pRL1383a as template (1:10 dilution of large-scale plasmid prep).
- 5uL Taq polymerase: Accusure(hot start, 68°C) and Red Taq
Note: the taq and water can be combined and aliquoted together as long as reaction is kept at 4°C.
- cold blocks
- pcr reaction tubes, with tops
- pipette and tips
| Duration of Time | T° 5 cycles | T° 30 cycles | Purpose |
|---|---|---|---|
| 10 minutes | 95°C | 95°C | heat activate taq |
| 30 seconds | 95°C | 95°C | denaturation |
| 30 seconds | 48.1°C, 53.4°C, 57.9°C | 59°C, 60.6°C, 61.9°C | annealing |
| 5 minutes | 68°C | 68°C | extension (1.5 minutes per kb, go with longest) |
| 10 minutes | 68°C | 68°C | finishing the extension |
| infinity | 4°C | 4°C | (until you remove product) |
Materials for the gel
- 1% agarose gel
- 10ug/mL Ethidium Bromide
- running apparatus
- gel running conditions: 95V for ~1 hour
Results
| Lane | Contents | Description | Predicted annealingT°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | ok | ||
| 2 | omega (aadA)#1 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 3 | oriV #1 | (+), ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 4 | mob #1 | smear | 53.5, 48 | 62, 69.8 |
| 5 | rep #1 | smear, low kb | 53.9, 57.2 | 59.1, 73.5 |
| 6 | (-)control | all clear | ||
| 7 | (+) control | bright band, ~0.5kb | ||
| 8 | omega #2 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 9 | oriV #2 | (+) ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 10 | mob #2 | nothing, slight smear | 53.5, 48 | 62, 69.8 |
| Lane | Contents | Description | Predicted annealing T°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | no ladder | ||
| 2 | rep#2 | light smear | 53.9, 57.2 | 59.1, 73.5 |
| 3 | (-)control | all clear | ||
| 4 | (+)control | bright band ?0.5kb | ||
| 5 | omega #3 | bright band, ?1kb | 54.4,50.4 | 60.7,69.5 |
| 6 | oriV #3 | bright band, 0.5kb? | 51.3, 53 | 60.9, 71.2 |
| 7 | mob | light band, some smear, 3kb? | 53.5, 48 | 62, 69.8 |
| 8 | rep #3 | band in low kb | 53.9, 57.2 | 59.1, 73.5 |
| 9 | (-) control | all clear | ||
| 10 | (+) control | bright band, 0.5kb? |
Discussion
- The gels are very warped. I did not let them dry long enough before I poured the running buffer,so some products did not run correctly therefore this experiment will be repeated on Monday when the necessary reagents are delivered.
- I mixed up the temperature gradient!!! I need to be more careful about labeling my reactions.
- The rep protein did not come out at all and the mob region only came out at one temperature, so these must be PCRed again to determine the correct annealing temperature.
Follow-up Experiments
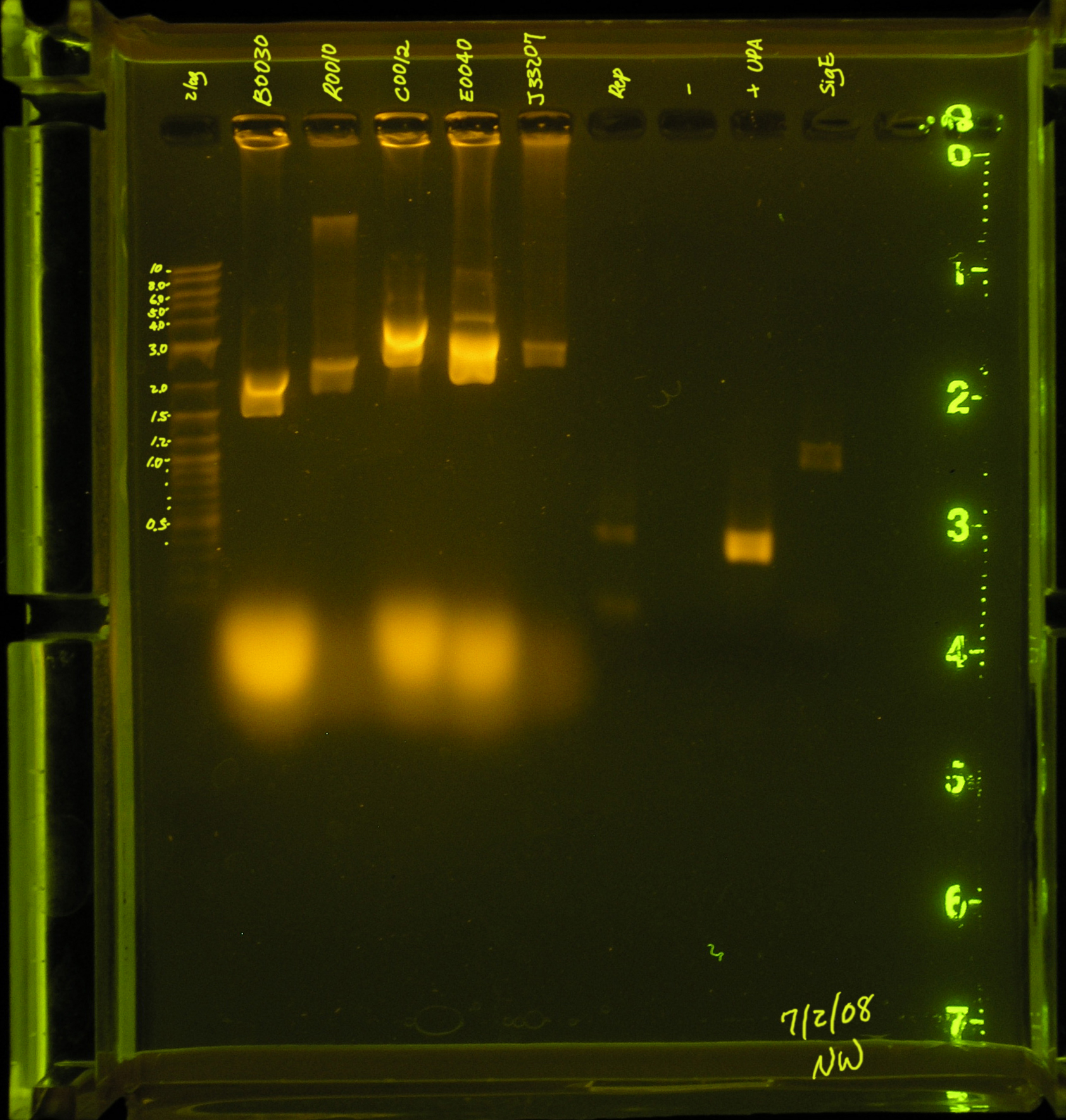
7/2/08
- A second attempt was made to PCR the rep region. An annealing temperature of 53.4° was used for the first 5 cycles and annealing temperature 60.6 was used for the subsequent 30 cycles.
- Results: There are 2 faint bands in the low kb region. This is probably primers.
- Discussion: The rep region did not amplify under these conditions. There were several suggestions made by the professors that I will try this weekend.
7/4/08</stong>
- Materials:
- 5uL Red Taq (per reaction)
- 3.5uL nanopure water (per reaction)
- 1uL 10uM f/r rep region primers (per reaction)
- 1uL of either 1:100, 1:500, 1:1000 dilution pRL1383a as template (1:10 dilution of large-scale plasmid prep)
- 5uL accusure (for one reaction to test the dilution with this enzyme)
| Duration of Time | T° 5 cycles | T° 30 cycles | Purpose |
|---|---|---|---|
| 10 minutes | 95°C | 95°C | heat activate taq |
| 30 seconds | 95°C | 95°C | denaturation |
| 30 seconds | 54 | 59 | annealing |
| 5 minutes | 68°C | 68°C | extension (1.5 minutes per kb, go with longest) |
| 10 minutes | 68°C | 68°C | finishing the extension |
| infinity | 4°C | 4°C | (until you remove product) |
Insanity is doing the same thing over and over again and expecting different results. - Albert Einstein
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"