|
_DNA-Origami
Introduction
Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.
In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances.
One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.
As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system.
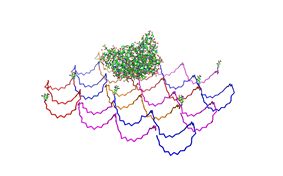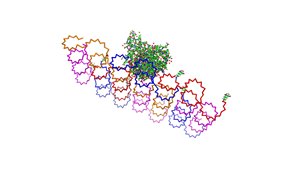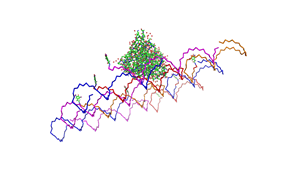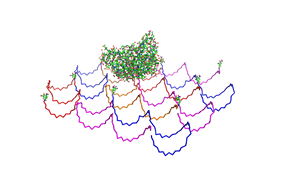 File:Freiburg2008 NIP arrangement schematical.jpg
File:Freiburg2008 NIP arrangement schematical.jpg
Literature
- Paul W. K. Rothemund: Nature 440, 297-302 (16 March 2006)
Methods
Phage DNA
Cell culture
50 ml DYT-Medium, 50µl tetracycline (TET; 25 mg/ ml) and ER2738-cells
were shaken over night at 37°C. The overnight culture was diluted with
DYT to OD600=0.1 and shaken at 37°C until the culture had an OD600
around 0.4. Each 50 ml of cell culture were inoculated with 5 µl
M13mp18 phage and shaken for 4 h at 37°C.
Isolation of M13mp18 phage from cell culture
PEG/NaCl was used to precipitate the phages.
First precipitation
Each 50 ml of cell culture were centrifuged at 5000 g for 20 min. While
the cell were centrifuged 1/7 volume of the supernatant PEG/NaCl (about
7ml/50ml) was put in a new falcon tube. The phages stay in the
supernatant therefore the supernatant was carefully decanted to the
PEG/NaCl and mixed gently by inverting the tube. The mixture (Solution
1) was left overnight at 4°C.
Solution 1 was centrifuged at 5000 g for 20 min. Because the phage stay
in the pellet, the supernatant was removed and the pellet was
resuspended in 2 ml TBS-Buffer (Solution 2). Solution 2 was put in a
1,5 ml Eppendorf tube and centrifuged (13200 rpm, 10 min). After the
centrifugation the phages stay in the supernatant.
Second precipitation
170 µl PEG/NaCl(~ 1/6 volume of supernatant) were put in a Eppendorf
tube. Supernatant was carefully decanted to the PEG/NaCl and mixed
gently by inverting the tube. The mixture (Solution 3) was left for 1 h
on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.
Define phage titers
The absorption of Solution 3 was measured on a Jasco V-550 UV/VIS
spectrometer at 269 nm.
Phage titer was calculated as follows:
Phage DNA =
((A269-A360) * 6 * 10^16 * dilution factor) / (number of bases in the
phage genom = 7249 bp)
Isolation of the phage DNA
The phage DNA was isolated with QIAprep Spin M13-Kit (50) from QIAGEN
(Cat.No: 27704).
DNA-concentration was quantified by Nano-drop photometer.
Origami
Produce
the Origami
To produce the Origami we mixed each the M13mp18 DNA with the oligos,
water and TEA/MgAcetat (end concentration =12.5mM).
<o:p></o:p>
<o:p> TABELLE 1</o:p>
<o:p>TABELLE 2</o:p>
Various samples were
produced. For the sample with a ratio of 1:20 (DNA:Oligo) we used 4 nM
DNA and 80 nM of oligos. The Origami were produced in a eppendorf
Mastercycler personal. Therefore they were heated up to 95°C for 7 min
and slowly cooled down (0.3°C/s) to 20°C.
Different sample were made:
Sample 1:20, 1:10 and 1:5 without NIP, oligos without NIP were used
Sample 1:20, 1:10 and 1:5 with NIP, all of the 7 oligos with NIP were
used -> origami with 7NIP
Sample 1:5 with NIP and fluorophor, all of the 7 oligos with NIP and
the 2 oligos with the Alexa 488 were used.
|  "
"

