Team:Cambridge/Voltage/Lab Work
From 2008.igem.org
15th July
Conference in Edinburgh
Meet with the other UK iGEM teams.
16th July
Conference in Edinburgh
Presentation of projects to the other teams and to the attendees of the SynBioStandards network meeting.
17th July
Progress
-Ordered mutant E.coli chassis to test: Kch-, KdpD-, KdpE-, KdpF-E-
-Designed two plasmids:
-Sourced V.harveyi BB170 culture to obtain glutamate-gated potassium channel gene
-Modelled protein structure
-Designed primers for extraction of Kdp Operon from E.coli and Glu Gated channel gene from V.harveyi
-Decided on oxygen electrode for small voltage change measurements
-Extracted BioBricks from registry to use in plasmids:
- Promoter BBa_J23100
- RBS BBa_B0030
- Terminators BBa_B1002 and BBa_B1006
18th July
Wet Work
There was no growth on the plates spread yesterday. There has been a few adjustments to the protocol.
Biobrick Extraction
| Class | Registry | Part Kit Location | Tube Label |
|---|---|---|---|
| Promoter | BBa_J23100 | 2008 1002-10E | F |
| RBS | BBa_B0030 | 2008 1002-5G | G |
| Stop | BBa_B1002 | 2008 1016-8A | H |
| Stop | BBa_B1006 | 2008 1016-8D | I |
| Promoter | BBa_J23113 | 2008 1002-12B | J |
| Promoter | BBa_J23114 | 2008 1002-12C | K |
| Control | pUC19 | L |
Bigger, better, faster, stronger
We found we could not extract enough DNA from the registry, so we are upgrading the extraction to the "bigger, better, faster, stronger" method.
Warm 50μL of EB in Eppendorf tubes at 50°C and add 4 punched spots. Keep it warming at 50°C for 20mins and spin down for 3 minutes at 15,000 g. Warm agin for 10mins and spin down again for 3mins. Pipette off the liquid which should have the DNA in. We then confirmed with PCR.
Each sample except the control was then put through 34 cycles of PCR.
The following were added to eppendorf tubes:
5μL of DNA in EB buffer
2.5μL of each primer for Biobrick vectors
25μL of Finnzymes mastermix
15μL of sterile distilled water
And all 50μL were then run through the PCR reaction and 17μL of product run on an E-Gel with 3μL dye.
The results gave large amounts of DNA in each lane for all Biobricks except F. The original extracts of G,H,I,J,K and L were then used for the following transformations.
To chilled tubes add 5μL of DNA + EB solution to 5μL of Cells ( Chemically competent Top 10 )
Ice for 30 minutes Heat shock at 42°C for 60 seconds Ice for 2 minutes Add 500μL of SOC Incubate at 37°C overnight
Prepare Agar plates : Add 200μL of Amp (100mg/mL) into 200mL of LA ( Final conc. of 100μg/mL ) and add 100μL of Amp (100mg/mL) into 200mL of LA ( Final conc. of 50μg/mL ) Plate neat samples of G,H,I,J,K,L onto both types of plate and incubate overnight.
21st July
Wet Work
Aims for Today
The results of yesterday's modified protocol still yielded no growth on the plates. Using yesterday's PCR product our aims were:
- Cut promoters with spe1
- Cut RBS with Xba1
- Ligate these together and perform further PCR amplification of linear fragments.
- Cut first 'stop' with spe1
- Cut second 'stop with xba1
- Ligate these together and perform further PCR amplification of linear fragments.
- Finally, put all the above elements into a vector plasmid.
- End of day: Aims completed
Length
- Promoter = 35 bases
- RBS = 16 bases
- Stop (B1002) = 35 bases and Stop (B1006) = 39 bases
22nd July
Wet Work
- Inoculated 10mL LB with E.coli strain MG1655
- Inoculated 10mL LB with E.coli strain TOP10 transformed with BioBrick J5311
16 LA plates were poured with 8 having 25μg/mL Kanamycin, ready for mutant strains to be grown on.
The gel extracts from the ligation of primer and rbs were extracted using Zymoclean gel extraction kit. Hopefully these now contain linear DNA of BBa_J23113 and BBa_B0030, and BBa_J23114 and BBa_B0030.
BioBricks J45996, J45995 and J45250 were also extracted from the registry, using the "bigger, better, faster, stronger, method", in order to provide template to extracted the OsmY promoter.
23rd July
Wet Work
- Recovered Biobrick plasmid YFP from previous culture.
- 1.6 ml of cells according to Zippy Plasmid Miniprep Kit protocol (60μL Elution Buffer used)
- Added 20ml LB to 10ml innoculation of E Coli MG1655 (see 22/07/08) to increase volume in preparation for experiments tomorrow on K+ tolerance in media.
24th July
Double Terminator
The double terminator was made from BBa_B1006 and BBa_B1002, combined together.
First the two Biobricks were cut using the fast digest enzymes, at either Xba or Spe. They were then run on an agarose gel, cut out and extracted, using Zymoclean gel extraction. The 2006 was not cut succesfully with Spe, so was not extracted. The samples were then ligated, using fast ligation, such that 1002-1002 and 1002-1006 were hopefully produced. This was done by combining the 1002 cut with Spe with the 1002 cut with Xba and the 1006 cut with Xba. These were then run on an agarose gel and cut out, then frozen overnight.
25th July
Double Terminator
The DNA was extracted from lanes 3&4 of the gel and extracted using the Zyppy Gel Extraction kit. The extracted DNA was then PCRed using 34 cycles, and then 10μL was frozen at -20°C. 8μL of the PCR product was cut with both EcoR1 and Pst1, as was 5μL of the death plasmid, PSB4C5, using the fast digest protocol. The gene was then ligated into the plasmid backbone, using the fast ligation kit, from Finnzymes.
500μL of normal TOP10 cells were made competent, and then transformed with the 4μL of the new plasmid, and grown on chloramphenicol overnight. 1μL of Puc19 was used as a control.
K+ assay for E.coli
Solutions of potassium chlotide were made up from 0mM to 500mM. 100μL of cells, 100μL of 10x KCl solution and 800μL of SDW were mixed and the OD600 measured every 30 mins, after agitation to ensure homogenaity. After 4 hours 800μL of the cell solution were spun down and resuspended in 1mL SDW, lysed using the freeze-thaw method, and the K+ concentration measured using the Flame Photometer.
28th July
The flame photometer was also calibrated using μM concentrations of potassium.
The mutants were streaked out from the innoculation discs onto both kanamycin and non antibiotic plates for the resistant strains, and just onto the normal plates for non-resistant strains. They were then incubated overnight at 37°C.
Mutant Plates
29th July
- Mutant E.coli strains had grown on plates.
- Transferred mutants to liquid culture.
- Primers arrived! Prepped as described in Protocols page
- PCR started with OsmY (from Biobrick plasmids) and KDP (from E. Coli MG1655 Cell Culture)
- E-Gel (SYBR) to confirm OsmY
- E-Gel (EtBr) to confirm Kdp and OsmY
30th July
Competence test
- Transformed PUK plasmid and YFP plasmid into mutant E.coli strains.
osmY and Kdp
- digested PCR products, and death gene vector with EcoR1 and Pst1
- ligated new BioBricks into vector
- Transformed into competent TOP10 cells
- Plate out onto Chloramphenicol plates
31st July
Mutants
- Mutant strains were grown in different potassium levels: 1mM, 50mM and 200mM.
- Growth curves were plotted, using OD600 readings every 30 minutes for 3 hours.
Kdp
- Ligated Kdp into death vector PSB4C5 again.
- More PCR of gene from MG1655.
OsmY
- TOP10 containing osmY-containing plasmid were grown in LB.
1st August
Mutants
- The OD600 of the mutants grown in different K+ concentrations was taken again.
osmY
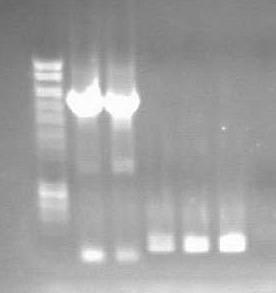
- Plasmid mini prep was done.
- Gel was run, showing clear band at around 450bp, indicating osmY with BioBrick primer prefix and suffix.
- Glycerol stocks were made of the TOP10 cultures with the osmY-containing plasmid.
- More LB was added to the TOP10 cultures and they were put in the incubator.
Plasmid
- PSB4C5 was extracted from DB31, into 100μl SDW - final conc. 106.5ng/μl.
4th August
Biobricks
- The liagtion reaction from last week was drop dialised and then 5μL used to transform highly competent TOP10, before plating on chloramphenicol plates.
- The Biobrick combination of promoter-RBS were then cut with Pst1 and EcoR1 and ligated into pSB4C5 and then transformed into TOP10 and incubated overnight at room temperature in 600μL SOC.
5th August
Kdp?
- There were transformants from yesterday's transformation
- After Biobrick PCR the gel showed short fragments, suggesting self ligation of the vector.
- Repeated digest and ligation protocol from the weekend, with EcoR1 and Pst1 instead of Xba1 and Spe.
Biobricks
- Extracted two GFP-only sequences (J04630 and I714062) from the registry, for use as reporters for osmY.
- PCR on these and the RBS B0030. Ran E-gel to check PCR.
- Cut osmY plasmid with Spe, and RBS B0030 with Xba, and ligated the two.
- Transformed cells with this construct. Then realised this was silly.
 "
"